Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion
- PMID: 38398085
- PMCID: PMC10887418
- DOI: 10.3390/cancers16040694
Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion
Abstract
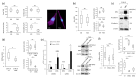
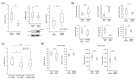
The intrinsic biomechanical properties of cancer cells remain poorly understood. To decipher whether cell stiffness modulation could increase melanoma cells' invasive capacity, we performed both in vitro and in vivo experiments exploring cell stiffness by atomic force microscopy (AFM). We correlated stiffness properties with cell morphology adaptation and the molecular mechanisms underlying epithelial-to-mesenchymal (EMT)-like phenotype switching. We found that melanoma cell stiffness reduction was systematically associated with the acquisition of invasive properties in cutaneous melanoma cell lines, human skin reconstructs, and Medaka fish developing spontaneous MAP-kinase-induced melanomas. We observed a systematic correlation of stiffness modulation with cell morphological changes towards mesenchymal characteristic gains. We accordingly found that inducing melanoma EMT switching by overexpressing the ZEB1 transcription factor, a major regulator of melanoma cell plasticity, was sufficient to decrease cell stiffness and transcriptionally induce tetraspanin-8-mediated dermal invasion. Moreover, ZEB1 expression correlated with Tspan8 expression in patient melanoma lesions. Our data suggest that intrinsic cell stiffness could be a highly relevant marker for human cutaneous melanoma development.
Keywords: EMT-TFs; biomechanics; melanoma; stiffness; tetraspanin 8.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Tsoi J., Robert L., Paraiso K., Galvan C., Sheu K.M., Lay J., Wong D.J.L., Atefi M., Shirazi R., Wang X., et al. Multi-Stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell. 2018;33:890–904.e5. doi: 10.1016/j.ccell.2018.03.017. - DOI - PMC - PubMed
-
- Hoek K.S., Schlegel N.C., Brafford P., Sucker A., Ugurel S., Kumar R., Weber B.L., Nathanson K.L., Phillips D.J., Herlyn M., et al. Metastatic Potential of Melanomas Defined by Specific Gene Expression Profiles with No BRAF Signature. Pigment. Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. - DOI - PubMed
-
- Widmer D.S., Cheng P.F., Eichhoff O.M., Belloni B.C., Zipser M.C., Schlegel N.C., Javelaud D., Mauviel A., Dummer R., Hoek K.S. Systematic Classification of Melanoma Cells by Phenotype-Specific Gene Expression Mapping. Pigment. Cell Melanoma Res. 2012;25:343–353. doi: 10.1111/j.1755-148X.2012.00986.x. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

