A Potential Prognostic Gene Signature Associated with p53-Dependent NTRK1 Activation and Increased Survival of Neuroblastoma Patients
- PMID: 38398114
- PMCID: PMC10886603
- DOI: 10.3390/cancers16040722
A Potential Prognostic Gene Signature Associated with p53-Dependent NTRK1 Activation and Increased Survival of Neuroblastoma Patients
Abstract
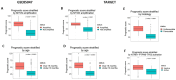
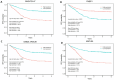
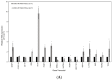
Neuroblastoma is the most common extracranial solid tumour in children, comprising close to 10% of childhood cancer-related deaths. We have demonstrated that activation of NTRK1 by TP53 repression of PTPN6 expression is significantly associated with favourable survival in neuroblastoma. The molecular mechanisms by which this activation elicits cell molecular changes need to be determined. This is critical to identify dependable biomarkers for the early detection and prognosis of tumours, and for the development of personalised treatment. In this investigation we have identified and validated a gene signature for the prognosis of neuroblastoma using genes differentially expressed upon activation of the NTRK1-PTPN6-TP53 module. A random survival forest model was used to construct a gene signature, which was then assessed across validation datasets using Kaplan-Meier analysis and ROC curves. The analysis demonstrated that high BASP1, CD9, DLG2, FNBP1, FRMD3, IL11RA, ISGF10, IQCE, KCNQ3, and TOX2, and low BSG/CD147, CCDC125, GABRB3, GNB2L1/RACK1 HAPLN4, HEBP2, and HSD17B12 expression was significantly associated with favourable patient event-free survival (EFS). The gene signature was associated with favourable tumour histology and NTRK1-PTPN6-TP53 module activation. Importantly, all genes were significantly associated with favourable EFS in an independent manner. Six of the signature genes, BSG/CD147, GNB2L1/RACK1, TXNDC5, FNPB1, B3GAT1, and IGSF10, play a role in cell differentiation. Our findings strongly suggest that the identified gene signature is a potential prognostic biomarker and therapeutic target for neuroblastoma patients and that it is associated with neuroblastoma cell differentiation through the activation of the NTRK1-PTPN6-TP53 module.
Keywords: NTRK1; biomarkers; gene signature; neuroblastoma; prognosis.
Conflict of interest statement
All the authors declare that there is no conflict of interest.
Figures


References
-
- Schleiermacher G., Mosseri V., London W.B., Maris J.M., Brodeur G.M., Attiyeh E., Haber M., Khan J., Nakagawara A., Speleman F., et al. Segmental Chromosomal Alterations Have Prognostic Impact in Neuroblastoma: A Report from the INRG Project. Br. J. Cancer. 2012;107:1418–1422. doi: 10.1038/bjc.2012.375. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

