A Comprehensive Study of the Effects by Sequence Truncation within Inverted Terminal Repeats (ITRs) on the Productivity, Genome Packaging, and Potency of AAV Vectors
- PMID: 38399714
- PMCID: PMC10892565
- DOI: 10.3390/microorganisms12020310
A Comprehensive Study of the Effects by Sequence Truncation within Inverted Terminal Repeats (ITRs) on the Productivity, Genome Packaging, and Potency of AAV Vectors
Abstract
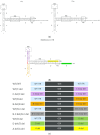
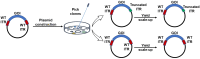
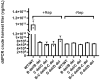
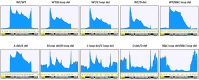
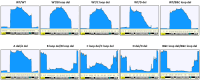
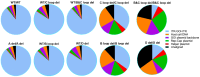
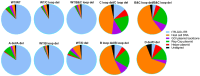
One of the primary challenges in working with adeno-associated virus (AAV) lies in the inherent instability of its inverted terminal repeats (ITRs), which play vital roles in AAV replication, encapsidation, and genome integration. ITRs contain a high GC content and palindromic structure, which occasionally results in truncations and mutations during plasmid amplification in bacterial cells. However, there is no thorough study on how these alterations in ITRs impact the ultimate AAV vector characteristics. To close this gap, we designed ITRs with common variations, including a single B, C, or D region deletion at one end, and dual deletions at both ends of the vector genome. These engineered ITR-carrying plasmids were utilized to generate AAV vectors in HEK293 cells. The crude and purified AAV samples were collected and analyzed for yield, capsid DNA-filled percentage, potency, and ITR integrity. The results show that a single deletion had minor impact on AAV productivity, packaging efficiency, and in vivo potency. However, deletions on both ends, except A, showed significant negative effects on the above characteristics. Our work revealed the role of ITR regions, A, B, C, and D for AAV production and DNA replication, and proposes a new strategy for the quality control of ITR-bearing plasmids and final AAV products.
Keywords: ITRs; nanopore sequencing; packaging; potency; productivity; rAAV.
Conflict of interest statement
The authors were employed by the company Alexion Pharmaceuticals. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
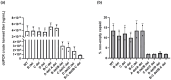
Similar articles
-
Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions.J Virol. 1996 Mar;70(3):1668-77. doi: 10.1128/JVI.70.3.1668-1677.1996. J Virol. 1996. PMID: 8627687 Free PMC article.
-
Impact of Inverted Terminal Repeat Integrity on rAAV8 Production Using the Baculovirus/Sf9 Cells System.Hum Gene Ther Methods. 2017 Oct;28(5):277-289. doi: 10.1089/hgtb.2016.133. Hum Gene Ther Methods. 2017. PMID: 28967288 Free PMC article.
-
Deletion of the B-B' and C-C' regions of inverted terminal repeats reduces rAAV productivity but increases transgene expression.Sci Rep. 2017 Jul 14;7(1):5432. doi: 10.1038/s41598-017-04054-4. Sci Rep. 2017. PMID: 28710345 Free PMC article.
-
A User's Guide to the Inverted Terminal Repeats of Adeno-Associated Virus.Hum Gene Ther Methods. 2019 Dec;30(6):206-213. doi: 10.1089/hgtb.2019.276. Hum Gene Ther Methods. 2019. PMID: 31752513 Review.
-
AAV- based vector improvements unrelated to capsid protein modification.Front Med (Lausanne). 2023 Feb 3;10:1106085. doi: 10.3389/fmed.2023.1106085. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36817775 Free PMC article. Review.
Cited by
-
Degradation and stable maintenance of adeno-associated virus inverted terminal repeats in E. coli.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1170. doi: 10.1093/nar/gkae1170. Nucleic Acids Res. 2025. PMID: 39657764 Free PMC article.
-
Consecutive Affinity and Ion-Exchange Chromatography for AAV9 Vectors Purification.Biomedicines. 2025 Feb 5;13(2):361. doi: 10.3390/biomedicines13020361. Biomedicines. 2025. PMID: 40002774 Free PMC article.
-
Improved Recombinant Adeno-Associated Viral Vector Production via Molecular Evolution of the Viral Rep Protein.Int J Mol Sci. 2025 Feb 4;26(3):1319. doi: 10.3390/ijms26031319. Int J Mol Sci. 2025. PMID: 39941089 Free PMC article.
-
Tracking Particle-Encapsulated DNA Across the Anion-Exchange Chromatography Fractions of Recombinant Adeno-Associated Virus Using Droplet Digital PCR and High-Throughput Sequencing.Biotechnol J. 2025 Jun;20(6):e70031. doi: 10.1002/biot.70031. Biotechnol J. 2025. PMID: 40490977 Free PMC article.
References
-
- Flotte T.R., Berns K.I. Adeno-associated viral vectors for gene therapy. Lab. Tech. Biochem. Mol. Biol. 2005;31:255–272. doi: 10.1016/S0075-7535(05)31001-1. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

