Optimizing the Multimerization Properties of Quinoline-Based Allosteric HIV-1 Integrase Inhibitors
- PMID: 38399977
- PMCID: PMC10892445
- DOI: 10.3390/v16020200
Optimizing the Multimerization Properties of Quinoline-Based Allosteric HIV-1 Integrase Inhibitors
Abstract
Allosteric HIV-1 Integrase (IN) Inhibitors or ALLINIs bind at the dimer interface of the IN, away from the enzymatic catalytic site, and disable viral replication by inducing over-multimerization of IN. Interestingly, these inhibitors are capable of impacting both the early and late stages of viral replication. To better understand the important binding features of multi-substituted quinoline-based ALLINIs, we have surveyed published studies on IN multimerization and antiviral properties of various substituted quinolines at the 4, 6, 7, and 8 positions. Here we show how the efficacy of these inhibitors can be modulated by the nature of the substitutions at those positions. These features not only improve the overall antiviral potencies of these compounds but also significantly shift the selectivity toward the viral maturation stage. Thus, to fully maximize the potency of ALLINIs, the interactions between the inhibitor and multiple IN subunits need to be simultaneously optimized.
Keywords: ALLINI; HIV; aberrant integrase multimerization; allosteric integrase inhibitor; integrase; quinoline; virus maturation.
Conflict of interest statement
The authors declare no conflicts of interest. The National Institutes of Health had no role in the writing of the manuscript; or in the decision to publish.
Figures
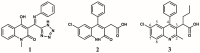
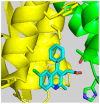
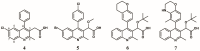
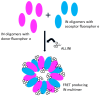
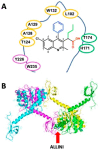
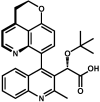
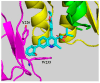
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

