The Potential of Usnic-Acid-Based Thiazolo-Thiophenes as Inhibitors of the Main Protease of SARS-CoV-2 Viruses
- PMID: 38399993
- PMCID: PMC10893357
- DOI: 10.3390/v16020215
The Potential of Usnic-Acid-Based Thiazolo-Thiophenes as Inhibitors of the Main Protease of SARS-CoV-2 Viruses
Abstract
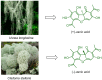
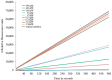
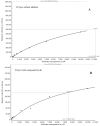
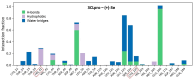
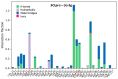
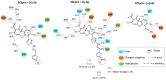
Although the COVID-19 pandemic caused by SARS-CoV-2 viruses is officially over, the search for new effective agents with activity against a wide range of coronaviruses is still an important task for medical chemists and virologists. We synthesized a series of thiazolo-thiophenes based on (+)- and (-)-usnic acid and studied their ability to inhibit the main protease of SARS-CoV-2. Substances containing unsubstituted thiophene groups or methyl- or bromo-substituted thiophene moieties showed moderate activity. Derivatives containing nitro substituents in the thiophene heterocycle-just as pure (+)- and (-)-usnic acids-showed no anti-3CLpro activity. Kinetic parameters of the most active compound, (+)-3e, were investigated, and molecular modeling of the possible interaction of the new thiazolo-thiophenes with the active site of the main protease was carried out. We evaluated the binding energies of the ligand and protein in a ligand-protein complex. Active compound (+)-3e was found to bind with minimum free energy; the binding of inactive compound (+)-3g is characterized by higher values of minimum free energy; the positioning of pure (+)-usnic acid proved to be unstable and is accompanied by the formation of intermolecular contacts with many amino acids of the catalytic binding site. Thus, the molecular dynamics results were consistent with the experimental data. In an in vitro antiviral assay against six strains (Wuhan, Delta, and four Omicron sublineages) of SARS-CoV-2, (+)-3e demonstrated pronounced antiviral activity against all the strains.
Keywords: 3CLpro; SARS-CoV-2; main protease; molecular modeling; usnic acid.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
(+)-Usnic Acid and Its Derivatives as Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses.Viruses. 2022 Sep 29;14(10):2154. doi: 10.3390/v14102154. Viruses. 2022. PMID: 36298709 Free PMC article.
-
Chetomin, a SARS-CoV-2 3C-like Protease (3CLpro) Inhibitor: In Silico Screening, Enzyme Docking, Molecular Dynamics and Pharmacokinetics Analysis.Viruses. 2023 Jan 15;15(1):250. doi: 10.3390/v15010250. Viruses. 2023. PMID: 36680290 Free PMC article.
-
Optimization Rules for SARS-CoV-2 Mpro Antivirals: Ensemble Docking and Exploration of the Coronavirus Protease Active Site.Viruses. 2020 Aug 26;12(9):942. doi: 10.3390/v12090942. Viruses. 2020. PMID: 32859008 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
Analyzing 3D structures of the SARS-CoV-2 main protease reveals structural features of ligand binding for COVID-19 drug discovery.Drug Discov Today. 2023 Oct;28(10):103727. doi: 10.1016/j.drudis.2023.103727. Epub 2023 Jul 27. Drug Discov Today. 2023. PMID: 37516343 Review.
Cited by
-
Exploring Thiophene Derivatives: Synthesis Strategies and Biological Significance.Med Chem. 2025;21(1):11-31. doi: 10.2174/0115734064326879240801043412. Med Chem. 2025. PMID: 39916435 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

