Unveiling CRESS DNA Virus Diversity in Oysters by Virome
- PMID: 38400004
- PMCID: PMC10892194
- DOI: 10.3390/v16020228
Unveiling CRESS DNA Virus Diversity in Oysters by Virome
Abstract
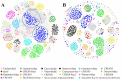
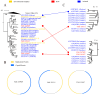
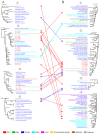
Oysters that filter feed can accumulate numerous pathogens, including viruses, which can serve as a valuable viral repository. As oyster farming becomes more prevalent, concerns are mounting about diseases that can harm both cultivated and wild oysters. Unfortunately, there is a lack of research on the viruses and other factors that can cause illness in shellfish. This means that it is harder to find ways to prevent these diseases and protect the oysters. This is part of a previously started project, the Dataset of Oyster Virome, in which we further study 30 almost complete genomes of oyster-associated CRESS DNA viruses. The replication-associated proteins and capsid proteins found in CRESS DNA viruses display varying evolutionary rates and frequently undergo recombination. Additionally, some CRESS DNA viruses have the capability for cross-species transmission. A plethora of unclassified CRESS DNA viruses are detectable in transcriptome libraries, exhibiting higher levels of transcriptional activity than those found in metagenome libraries. The study significantly enhances our understanding of the diversity of oyster-associated CRESS DNA viruses, emphasizing the widespread presence of CRESS DNA viruses in the natural environment and the substantial portion of CRESS DNA viruses that remain unidentified. This study's findings provide a basis for further research on the biological and ecological roles of viruses in oysters and their environment.
Keywords: CRESS DNA virus; Cap; Rep; oyster; phylogenetic tree; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Genetic Diversity and Characterization of Circular Replication (Rep)-Encoding Single-Stranded (CRESS) DNA Viruses.Microbiol Spectr. 2022 Dec 21;10(6):e0105722. doi: 10.1128/spectrum.01057-22. Epub 2022 Nov 8. Microbiol Spectr. 2022. PMID: 36346238 Free PMC article.
-
Unveiling the Virome of Wild Birds: Exploring CRESS-DNA Viral Dark Matter.Genome Biol Evol. 2024 Oct 9;16(10):evae206. doi: 10.1093/gbe/evae206. Genome Biol Evol. 2024. PMID: 39327897 Free PMC article.
-
Diversity of CRESS DNA Viruses in Squamates Recapitulates Hosts Dietary and Environmental Sources of Exposure.Microbiol Spectr. 2022 Jun 29;10(3):e0078022. doi: 10.1128/spectrum.00780-22. Epub 2022 May 26. Microbiol Spectr. 2022. PMID: 35616383 Free PMC article.
-
Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses With Small Genomes and a Diverse Host Range.Adv Virus Res. 2019;103:71-133. doi: 10.1016/bs.aivir.2018.10.001. Epub 2018 Dec 5. Adv Virus Res. 2019. PMID: 30635078 Review.
-
Counts and sequences, observations that continue to change our understanding of viruses in nature.J Microbiol. 2015 Mar;53(3):181-92. doi: 10.1007/s12275-015-5068-6. Epub 2015 Mar 3. J Microbiol. 2015. PMID: 25732739 Review.
Cited by
-
Identification and classification of the genomes of novel microviruses in poultry slaughterhouse.Front Microbiol. 2024 May 2;15:1393153. doi: 10.3389/fmicb.2024.1393153. eCollection 2024. Front Microbiol. 2024. PMID: 38756731 Free PMC article.
-
A comprehensive RNA virome identified in the oyster Magallana gigas reveals the intricate network of virus sharing between seawater and mollusks.Microbiome. 2024 Dec 20;12(1):263. doi: 10.1186/s40168-024-01967-x. Microbiome. 2024. PMID: 39707493 Free PMC article.
-
Exploring the recombinant evolution and hosts of crucivirus based on novel oyster-associated viruses.Front Microbiol. 2025 Feb 4;16:1454079. doi: 10.3389/fmicb.2025.1454079. eCollection 2025. Front Microbiol. 2025. PMID: 39967733 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

