Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1
- PMID: 38400023
- PMCID: PMC10892870
- DOI: 10.3390/v16020247
Porcine Sapovirus Protease Controls the Innate Immune Response and Targets TBK1
Abstract
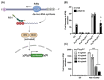
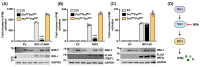
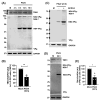
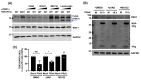
Human sapoviruses (HuSaVs) and noroviruses are considered the leading cause of acute gastroenteritis worldwide. While extensive research has focused on noroviruses, our understanding of sapoviruses (SaVs) and their interactions with the host's immune response remains limited. HuSaVs have been challenging to propagate in vitro, making the porcine sapovirus (PSaV) Cowden strain a valuable model for studying SaV pathogenesis. In this study we show, for the first time, that PSaV Cowden strain has mechanisms to evade the host's innate immune response. The virus 3C-like protease (NS6) inhibits type I IFN production by targeting TBK1. Catalytically active NS6, both during ectopic expression and during PSaV infection, targets TBK1 which is then led for rapid degradation by the proteasome. Moreover, deletion of TBK1 from porcine cells led to an increase in PSaV titres, emphasizing its role in regulating PSaV infection. Additionally, we successfully established PSaV infection in IPEC-J2 cells, an enterocytic cell line originating from the jejunum of a neonatal piglet. Overall, this study provides novel insights into PSaV evasion strategies, opening the way for future investigations into SaV-host interactions, and enabling the use of a new cell line model for PSaV research.
Keywords: IPEC-J2; NS6; TBK1; caliciviruses; innate immunity; protease; proteasomal degradation; sapovirus; type I IFN.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

