Validation of Candidate Host Cell Entry Factors for Bovine Herpes Virus Type-1 Based on a Genome-Wide CRISPR Knockout Screen
- PMID: 38400072
- PMCID: PMC10893506
- DOI: 10.3390/v16020297
Validation of Candidate Host Cell Entry Factors for Bovine Herpes Virus Type-1 Based on a Genome-Wide CRISPR Knockout Screen
Abstract
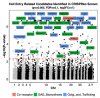
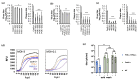
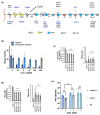
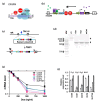
To identify host factors that affect Bovine Herpes Virus Type 1 (BoHV-1) infection we previously applied a genome wide CRISPR knockout screen targeting all bovine protein coding genes. By doing so we compiled a list of both pro-viral and anti-viral proteins involved in BoHV-1 replication. Here we provide further analysis of those that are potentially involved in viral entry into the host cell. We first generated single cell knockout clones deficient in some of the candidate genes for validation. We provide evidence that Polio Virus Receptor-related protein (PVRL2) serves as a receptor for BoHV-1, mediating more efficient entry than the previously identified Polio Virus Receptor (PVR). By knocking out two enzymes that catalyze HSPG chain elongation, HST2ST1 and GLCE, we further demonstrate the significance of HSPG in BoHV-1 entry. Another intriguing cluster of candidate genes, COG1, COG2 and COG4-7 encode six subunits of the Conserved Oligomeric Golgi (COG) complex. MDBK cells lacking COG6 produced fewer but bigger plaques compared to control cells, suggesting more efficient release of newly produced virions from these COG6 knockout cells, due to impaired HSPG biosynthesis. We further observed that viruses produced by the COG6 knockout cells consist of protein(s) with reduced N-glycosylation, potentially explaining their lower infectivity. To facilitate candidate validation, we also detailed a one-step multiplex CRISPR interference (CRISPRi) system, an orthogonal method to KO that enables quick and simultaneous deployment of three CRISPRs for efficient gene inactivation. Using CRISPR3i, we verified eight candidates that have been implicated in the synthesis of surface heparan sulfate proteoglycans (HSPGs). In summary, our experiments confirmed the two receptors PVR and PVRL2 for BoHV-1 entry into the host cell and other factors that affect this process, likely through the direct or indirect roles they play during HSPG synthesis and glycosylation of viral proteins.
Keywords: BoHV-1; COG complex; CRISPR/Cas9; Golgi apparatus; cell entry; heparan Sulfate; receptors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
GARP and EARP are required for efficient BoHV-1 replication as identified by a genome wide CRISPR knockout screen.PLoS Pathog. 2023 Dec 6;19(12):e1011822. doi: 10.1371/journal.ppat.1011822. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38055775 Free PMC article.
-
Conserved Oligomeric Golgi (COG) Complex Proteins Facilitate Orthopoxvirus Entry, Fusion and Spread.Viruses. 2020 Jun 30;12(7):707. doi: 10.3390/v12070707. Viruses. 2020. PMID: 32629851 Free PMC article.
-
A Genome-Wide CRISPR-Cas9 Screen Reveals the Requirement of Host Cell Sulfation for Schmallenberg Virus Infection.J Virol. 2020 Aug 17;94(17):e00752-20. doi: 10.1128/JVI.00752-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32522852 Free PMC article.
-
Heparan Sulfate Proteoglycans in Viral Infection and Treatment: A Special Focus on SARS-CoV-2.Int J Mol Sci. 2021 Jun 18;22(12):6574. doi: 10.3390/ijms22126574. Int J Mol Sci. 2021. PMID: 34207476 Free PMC article. Review.
-
Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias?Viruses. 2019 Jul 1;11(7):596. doi: 10.3390/v11070596. Viruses. 2019. PMID: 31266258 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

