Epigenetic modulation of myeloid cell functions in HIV and SARS-CoV-2 infection
- PMID: 38400997
- PMCID: PMC10894183
- DOI: 10.1007/s11033-024-09266-2
Epigenetic modulation of myeloid cell functions in HIV and SARS-CoV-2 infection
Abstract
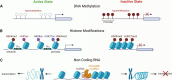
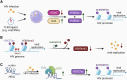
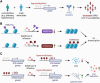
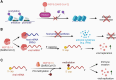
Myeloid cells play a vital role in innate immune responses as they recognize and phagocytose pathogens like viruses, present antigens, produce cytokines, recruit other immune cells to combat infections, and contribute to the attenuation of immune responses to restore homeostasis. Signal integration by pathogen recognition receptors enables myeloid cells to adapt their functions by a network of transcription factors and chromatin remodelers. This review provides a brief overview of the subtypes of myeloid cells and the main epigenetic regulation mechanisms. Special focus is placed on the epigenomic alterations in viral nucleic acids of HIV and SARS-CoV-2 along with the epigenetic changes in the host's myeloid cell compartment. These changes are important as they lead to immune suppression and promote the progression of the disease. Finally, we highlight some promising examples of 'epidrugs' that modulate the epigenome of immune cells and could be used as therapeutics for viral infections.
Keywords: AIDS; COVID-19; Epigenetics; HIV; Myeloid cells; SARS-CoV-2.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Modulation of host epigenome by coronavirus infections and developing treatment modalities for COVID-19 beyond genetics.Eur Rev Med Pharmacol Sci. 2021 Oct;25(19):5947-5964. doi: 10.26355/eurrev_202110_26872. Eur Rev Med Pharmacol Sci. 2021. PMID: 34661254 Review.
-
Harnessing Epigenetics: Innovative Approaches in Diagnosing and Combating Viral Acute Respiratory Infections.Pathogens. 2025 Feb 1;14(2):129. doi: 10.3390/pathogens14020129. Pathogens. 2025. PMID: 40005506 Free PMC article. Review.
-
Involvement of epigenetics in affecting host immunity during SARS-CoV-2 infection.Biochim Biophys Acta Mol Basis Dis. 2023 Mar;1869(3):166634. doi: 10.1016/j.bbadis.2022.166634. Epub 2022 Dec 26. Biochim Biophys Acta Mol Basis Dis. 2023. PMID: 36577469 Free PMC article. Review.
-
Epigenetic Lens to Visualize the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Infection in COVID-19 Pandemic.Front Genet. 2021 Mar 22;12:581726. doi: 10.3389/fgene.2021.581726. eCollection 2021. Front Genet. 2021. PMID: 33828579 Free PMC article. Review.
-
SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2.Immunity. 2021 Jun 8;54(6):1304-1319.e9. doi: 10.1016/j.immuni.2021.05.006. Epub 2021 May 9. Immunity. 2021. PMID: 34048708 Free PMC article.
Cited by
-
Is There such a Thing as Post-Viral Depression?: Implications for Precision Medicine.Biomol Ther (Seoul). 2024 Nov 1;32(6):659-684. doi: 10.4062/biomolther.2024.170. Epub 2024 Oct 21. Biomol Ther (Seoul). 2024. PMID: 39428555 Free PMC article. Review.
-
An Epigenetic Locus Associated with Loss of Smell in COVID-19.Diagnostics (Basel). 2024 Dec 15;14(24):2823. doi: 10.3390/diagnostics14242823. Diagnostics (Basel). 2024. PMID: 39767184 Free PMC article.
-
HIV-Induced Sialoglycans on Infected Cells Promote Immune Evasion from Myeloid Cell-Mediated Killing.bioRxiv [Preprint]. 2025 Feb 27:2025.02.21.639489. doi: 10.1101/2025.02.21.639489. bioRxiv. 2025. PMID: 40060686 Free PMC article. Preprint.
References
-
- Biswas SK, Mantovani A (2010) Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 11:889–896. 10.1038/ni.1937 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

