Enhancing grapevine breeding efficiency through genomic prediction and selection index
- PMID: 38401528
- PMCID: PMC10989862
- DOI: 10.1093/g3journal/jkae038
Enhancing grapevine breeding efficiency through genomic prediction and selection index
Abstract
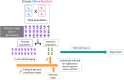
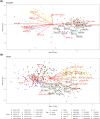
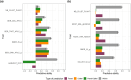
Grapevine (Vitis vinifera) breeding reaches a critical point. New cultivars are released every year with resistance to powdery and downy mildews. However, the traditional process remains time-consuming, taking 20-25 years, and demands the evaluation of new traits to enhance grapevine adaptation to climate change. Until now, the selection process has relied on phenotypic data and a limited number of molecular markers for simple genetic traits such as resistance to pathogens, without a clearly defined ideotype, and was carried out on a large scale. To accelerate the breeding process and address these challenges, we investigated the use of genomic prediction, a methodology using molecular markers to predict genotypic values. In our study, we focused on 2 existing grapevine breeding programs: Rosé wine and Cognac production. In these programs, several families were created through crosses of emblematic and interspecific resistant varieties to powdery and downy mildews. Thirty traits were evaluated for each program, using 2 genomic prediction methods: Genomic Best Linear Unbiased Predictor and Least Absolute Shrinkage Selection Operator. The results revealed substantial variability in predictive abilities across traits, ranging from 0 to 0.9. These discrepancies could be attributed to factors such as trait heritability and trait characteristics. Moreover, we explored the potential of across-population genomic prediction by leveraging other grapevine populations as training sets. Integrating genomic prediction allowed us to identify superior individuals for each program, using multivariate selection index method. The ideotype for each breeding program was defined collaboratively with representatives from the wine-growing sector.
Keywords: Cognac; GenPred; Genomic Prediction; Rosé; Shared Data Resource; genomic prediction; genomic selection; grapevine; ideotype; plant breeding; selection index.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest. The author(s) declare no conflicts of interest.
Figures


References
-
- Andrews S. 2016. FastQC a quality control tool for high throughput sequence data [WWW Document]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [accessed 2023 Jun 12].
-
- Bates D, Mächler M, Bolker B, Walker S. 2014. Fitting linear mixed-effects models using lme4. arXiv:1406.5823, preprint: not peer reviewed.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
