Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
- PMID: 38402281
- PMCID: PMC10894238
- DOI: 10.1038/s41467-024-45969-7
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Abstract
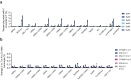
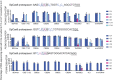
TadA-derived cytosine base editors (TadCBEs) enable programmable C•G-to-T•A editing while retaining the small size, high on-target activity, and low off-target activity of TadA deaminases. Existing TadCBEs, however, exhibit residual A•T-to-G•C editing at certain positions and lower editing efficiencies at some sequence contexts and with non-SpCas9 targeting domains. To address these limitations, we use phage-assisted evolution to evolve CBE6s from a TadA-mediated dual cytosine and adenine base editor, discovering mutations at N46 and Y73 in TadA that prevent A•T-to-G•C editing and improve C•G-to-T•A editing with expanded sequence-context compatibility, respectively. In E. coli, CBE6 variants offer high C•G-to-T•A editing and no detected A•T-to-G•C editing in any sequence context. In human cells, CBE6 variants exhibit broad Cas domain compatibility and retain low off-target editing despite exceeding BE4max and previous TadCBEs in on-target editing efficiency. Finally, we show that the high selectivity of CBE6 variants is well-suited for therapeutically relevant stop codon installation without creating unwanted missense mutations from residual A•T-to-G•C editing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare competing financial interests: The Broad Institute has filed a patent application on behalf of E.Z., M.E.N., and D.R.L on the base editors developed in this study. D.R.L. is a consultant for Prime Medicine, Beam Therapeutics, Pairwise Plants, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver agents for genome editing, epigenome engineering, or PACE, and owns equity in these companies. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

