Delivery of loaded MR1 monomer results in efficient ligand exchange to host MR1 and subsequent MR1T cell activation
- PMID: 38402309
- PMCID: PMC10894271
- DOI: 10.1038/s42003-024-05912-4
Delivery of loaded MR1 monomer results in efficient ligand exchange to host MR1 and subsequent MR1T cell activation
Abstract
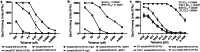
MR1-restricted T cells have been implicated in microbial infections, sterile inflammation, wound healing and cancer. Similar to other antigen presentation molecules, evidence supports multiple, complementary MR1 antigen presentation pathways. To investigate ligand exchange pathways for MR1, we used MR1 monomers and tetramers loaded with 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU) to deliver the antigen. Using MR1-deficient cells reconstituted with wild-type MR1 or MR1 molecules that cannot bind 5-OP-RU, we show that presentation of monomer-delivered 5-OP-RU is dependent on cellular MR1 and requires the transfer of ligand from the soluble molecule onto MR1 expressed by the antigen presenting cell. This mode of antigen delivery strengthens the evidence for post-ER ligand exchange pathways for MR1, which could represent an important avenue by which MR1 acquires antigens derived from endocytosed pathogens.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

