Bioframe: operations on genomic intervals in Pandas dataframes
- PMID: 38402507
- PMCID: PMC10903647
- DOI: 10.1093/bioinformatics/btae088
Bioframe: operations on genomic intervals in Pandas dataframes
Abstract
Motivation: Genomic intervals are one of the most prevalent data structures in computational genome biology, and used to represent features ranging from genes, to DNA binding sites, to disease variants. Operations on genomic intervals provide a language for asking questions about relationships between features. While there are excellent interval arithmetic tools for the command line, they are not smoothly integrated into Python, one of the most popular general-purpose computational and visualization environments.
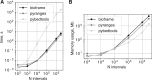
Results: Bioframe is a library to enable flexible and performant operations on genomic interval dataframes in Python. Bioframe extends the Python data science stack to use cases for computational genome biology by building directly on top of two of the most commonly-used Python libraries, NumPy and Pandas. The bioframe API enables flexible name and column orders, and decouples operations from data formats to avoid unnecessary conversions, a common scourge for bioinformaticians. Bioframe achieves these goals while maintaining high performance and a rich set of features.
Availability and implementation: Bioframe is open-source under MIT license, cross-platform, and can be installed from the Python Package Index. The source code is maintained by Open2C on GitHub at https://github.com/open2c/bioframe.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
No competing interest is declared.
Figures
Similar articles
-
PyRanges: efficient comparison of genomic intervals in Python.Bioinformatics. 2020 Feb 1;36(3):918-919. doi: 10.1093/bioinformatics/btz615. Bioinformatics. 2020. PMID: 31373614
-
Pygenomics: manipulating genomic intervals and data files in Python.Bioinformatics. 2023 Jun 1;39(6):btad346. doi: 10.1093/bioinformatics/btad346. Bioinformatics. 2023. PMID: 37228014 Free PMC article.
-
Gos: a declarative library for interactive genomics visualization in Python.Bioinformatics. 2023 Jan 1;39(1):btad050. doi: 10.1093/bioinformatics/btad050. Bioinformatics. 2023. PMID: 36688709 Free PMC article.
-
Explore, edit and leverage genomic annotations using Python GTF toolkit.Bioinformatics. 2019 Sep 15;35(18):3487-3488. doi: 10.1093/bioinformatics/btz116. Bioinformatics. 2019. PMID: 30768152
-
Pybedtools: a flexible Python library for manipulating genomic datasets and annotations.Bioinformatics. 2011 Dec 15;27(24):3423-4. doi: 10.1093/bioinformatics/btr539. Epub 2011 Sep 23. Bioinformatics. 2011. PMID: 21949271 Free PMC article.
Cited by
-
Efficient Hi-C inversion facilitates chromatin folding mechanism discovery and structure prediction.bioRxiv [Preprint]. 2023 Jul 21:2023.03.17.533194. doi: 10.1101/2023.03.17.533194. bioRxiv. 2023. Update in: Biophys J. 2023 Sep 5;122(17):3425-3438. doi: 10.1016/j.bpj.2023.07.017. PMID: 36993500 Free PMC article. Updated. Preprint.
-
ZNF143 is a transcriptional regulator of nuclear-encoded mitochondrial genes that acts independently of looping and CTCF.Mol Cell. 2025 Jan 2;85(1):24-41.e11. doi: 10.1016/j.molcel.2024.11.031. Epub 2024 Dec 20. Mol Cell. 2025. PMID: 39708805 Free PMC article.
-
Mitotic chromosomes are self-entangled and disentangle through a topoisomerase-II-dependent two-stage exit from mitosis.Mol Cell. 2024 Apr 18;84(8):1422-1441.e14. doi: 10.1016/j.molcel.2024.02.025. Epub 2024 Mar 22. Mol Cell. 2024. PMID: 38521067 Free PMC article.
-
Analysis-ready VCF at Biobank scale using Zarr.Gigascience. 2025 Jan 6;14:giaf049. doi: 10.1093/gigascience/giaf049. Gigascience. 2025. PMID: 40451243 Free PMC article.
-
Loss of multi-level 3D genome organization during breast cancer progression.bioRxiv [Preprint]. 2024 Aug 8:2023.11.26.568711. doi: 10.1101/2023.11.26.568711. bioRxiv. 2024. PMID: 38076897 Free PMC article. Preprint.
References
-
- Akalin A, Franke V, Vlahoviček K. et al. Genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 2015;31:1127–9. - PubMed
-
- den Bossche JV, Jordahl K, Fleischmann M. et al. geopandas/geopandas: v0.14.3. 2024. 10.5281/zenodo.2585848 - DOI
-
- Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng 2007;9:90–5.

