SEPN1-related myopathy depends on the oxidoreductase ERO1A and is druggable with the chemical chaperone TUDCA
- PMID: 38402623
- PMCID: PMC10982971
- DOI: 10.1016/j.xcrm.2024.101439
SEPN1-related myopathy depends on the oxidoreductase ERO1A and is druggable with the chemical chaperone TUDCA
Abstract
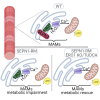
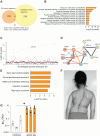
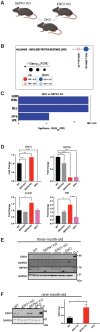
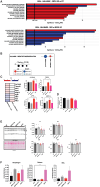
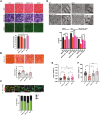
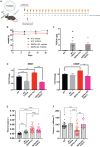
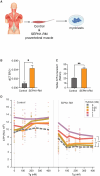
Selenoprotein N (SEPN1) is a protein of the endoplasmic reticulum (ER) whose inherited defects originate SEPN1-related myopathy (SEPN1-RM). Here, we identify an interaction between SEPN1 and the ER-stress-induced oxidoreductase ERO1A. SEPN1 and ERO1A, both enriched in mitochondria-associated membranes (MAMs), are involved in the redox regulation of proteins. ERO1A depletion in SEPN1 knockout cells restores ER redox, re-equilibrates short-range MAMs, and rescues mitochondrial bioenergetics. ERO1A knockout in a mouse background of SEPN1 loss blunts ER stress and improves multiple MAM functions, including Ca2+ levels and bioenergetics, thus reversing diaphragmatic weakness. The treatment of SEPN1 knockout mice with the ER stress inhibitor tauroursodeoxycholic acid (TUDCA) mirrors the results of ERO1A loss. Importantly, muscle biopsies from patients with SEPN1-RM exhibit ERO1A overexpression, and TUDCA-treated SEPN1-RM patient-derived primary myoblasts show improvement in bioenergetics. These findings point to ERO1A as a biomarker and a viable target for intervention and to TUDCA as a pharmacological treatment for SEPN1-RM.
Keywords: ER stress; ERO1; SEPN1; TUDCA; core myopathy; multi mini-core disease.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Chernorudskiy A., Varone E., Colombo S.F., Fumagalli S., Cagnotto A., Cattaneo A., Briens M., Baltzinger M., Kuhn L., Bachi A., et al. Selenoprotein N is an endoplasmic reticulum calcium sensor that links luminal calcium levels to a redox activity. Proc. Natl. Acad. Sci. USA. 2020;117:21288–21298. doi: 10.1073/pnas.2003847117. - DOI - PMC - PubMed
-
- Marino M., Stoilova T., Giorgi C., Bachi A., Cattaneo A., Auricchio A., Pinton P., Zito E. SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyper-oxidation by means of redox-regulating SERCA2 pump activity. Hum. Mol. Genet. 2015;24:1843–1855. - PubMed
-
- Pozzer D., Varone E., Chernorudskiy A., Schiarea S., Missiroli S., Giorgi C., Pinton P., Canato M., Germinario E., Nogara L., et al. A maladaptive ER stress response triggers dysfunction in highly active muscles of mice with SELENON loss. Redox Biol. 2019;20:354–366. doi: 10.1016/j.redox.2018.10.017. - DOI - PMC - PubMed
-
- Filipe A., Chernorudskiy A., Arbogast S., Varone E., Villar-Quiles R.N., Pozzer D., Moulin M., Fumagalli S., Cabet E., Dudhal S., et al. Defective endoplasmic reticulum-mitochondria contacts and bioenergetics in SEPN1-related myopathy. Cell Death Differ. 2021;28:123–138. doi: 10.1038/s41418-020-0587-z. - DOI - PMC - PubMed
-
- Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

