Epigenetic marks or not? The discovery of novel DNA modifications in eukaryotes
- PMID: 38403247
- PMCID: PMC11065753
- DOI: 10.1016/j.jbc.2024.106791
Epigenetic marks or not? The discovery of novel DNA modifications in eukaryotes
Abstract
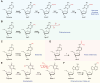
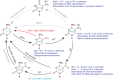
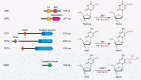
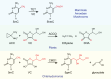
DNA modifications add another layer of complexity to the eukaryotic genome to regulate gene expression, playing critical roles as epigenetic marks. In eukaryotes, the study of DNA epigenetic modifications has been confined to 5mC and its derivatives for decades. However, rapid developing approaches have witnessed the expansion of DNA modification reservoirs during the past several years, including the identification of 6mA, 5gmC, 4mC, and 4acC in diverse organisms. However, whether these DNA modifications function as epigenetic marks requires careful consideration. In this review, we try to present a panorama of all the DNA epigenetic modifications in eukaryotes, emphasizing recent breakthroughs in the identification of novel DNA modifications. The characterization of their roles in transcriptional regulation as potential epigenetic marks is summarized. More importantly, the pathways for generating or eliminating these DNA modifications, as well as the proteins involved are comprehensively dissected. Furthermore, we briefly discuss the potential challenges and perspectives, which should be taken into account while investigating novel DNA modifications.
Keywords: 5-hydroxymethylcytosine (5hmC); 5-methylcytosine (5mC); DNA demethylation; DNA methylation; DNA modifications; N(6)-methyladenosine (6mA); TET dioxygenases; epigenetic marks; eukaryotes; transcriptional regulation.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
Similar articles
-
Modified Forms of Cytosine in Eukaryotes: DNA (De)methylation and Beyond.Methods Mol Biol. 2021;2198:3-13. doi: 10.1007/978-1-0716-0876-0_1. Methods Mol Biol. 2021. PMID: 32822018 Review.
-
Evidence for Noncytosine Epigenetic DNA Modifications in Multicellular Eukaryotes: An Overview.Methods Mol Biol. 2021;2198:15-25. doi: 10.1007/978-1-0716-0876-0_2. Methods Mol Biol. 2021. PMID: 32822019 Review.
-
Bacterial N4-methylcytosine as an epigenetic mark in eukaryotic DNA.Nat Commun. 2022 Feb 28;13(1):1072. doi: 10.1038/s41467-022-28471-w. Nat Commun. 2022. PMID: 35228526 Free PMC article.
-
N4-acetyldeoxycytosine DNA modification marks euchromatin regions in Arabidopsis thaliana.Genome Biol. 2022 Jan 3;23(1):5. doi: 10.1186/s13059-021-02578-7. Genome Biol. 2022. PMID: 34980211 Free PMC article.
-
Advances in mapping the epigenetic modifications of 5-methylcytosine (5mC), N6-methyladenine (6mA), and N4-methylcytosine (4mC).Biotechnol Bioeng. 2021 Nov;118(11):4204-4216. doi: 10.1002/bit.27911. Epub 2021 Aug 20. Biotechnol Bioeng. 2021. PMID: 34370308 Review.
Cited by
-
Decoding the genome and epigenome of avian Escherichia coli strains by R10.4.1 nanopore sequencing.Front Vet Sci. 2025 Mar 19;12:1541964. doi: 10.3389/fvets.2025.1541964. eCollection 2025. Front Vet Sci. 2025. PMID: 40177680 Free PMC article.
-
Synthesis of sensitive oligodeoxynucleotides containing acylated cytosine, adenine, and guanine nucleobases.DNA (Basel). 2025 Jun;5(2):25. doi: 10.3390/dna5020025. Epub 2025 May 9. DNA (Basel). 2025. PMID: 40709081 Free PMC article.
-
N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs.Int J Mol Sci. 2025 Feb 20;26(5):1812. doi: 10.3390/ijms26051812. Int J Mol Sci. 2025. PMID: 40076440 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

