FoxO1 promotes ovarian cancer by increasing transcription and METTL14-mediated m6A modification of SMC4
- PMID: 38403332
- PMCID: PMC11006996
- DOI: 10.1111/cas.16120
FoxO1 promotes ovarian cancer by increasing transcription and METTL14-mediated m6A modification of SMC4
Abstract
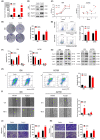
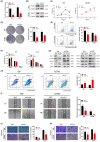
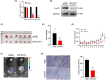
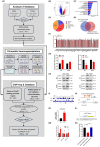
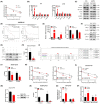
The transcription factor forkhead box protein O1 (FoxO1) is closely related to the occurrence and development of ovarian cancer (OC), however its role and molecular mechanisms remain unclear. Herein, we found that FoxO1 was highly expressed in clinical samples of OC patients and was significantly correlated with poor prognosis. FoxO1 knockdown inhibited the proliferation of OC cells in vitro and in vivo. ChIP-seq combined with GEPIA2 and Kaplan-Meier database analysis showed that structural maintenance of chromosome 4 (SMC4) is a downstream target of FoxO1, and FoxO1 promotes SMC4 transcription by binding to its -1400/-1390 bp promoter. The high expression of SMC4 significantly blocked the tumor inhibition effect of FoxO1 knockdown. Furtherly, FoxO1 increased SMC4 mRNA abundance by transcriptionally activating methyltransferase-like 14 (METTL14) and increasing SMC4 m6A methylation on its coding sequence region. The Cancer Genome Atlas dataset analysis confirmed a significant positive correlation between FoxO1, SMC4, and METTL14 expression in OC. In summary, this study revealed the molecular mechanisms of FoxO1 regulating SMC4 and established a clinical link between the expression of FoxO1/METTL14/SMC4 in the occurrence of OC, thus providing a potential diagnostic target and therapeutic strategy.
Keywords: FoxO1; METTL14; SMC4; m6A modification; ovarian cancer progression.
© 2024 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
SMC4 knockdown inhibits malignant biological behaviors of endometrial cancer cells by regulation of FoxO1 activity.Arch Biochem Biophys. 2021 Nov 15;712:109026. doi: 10.1016/j.abb.2021.109026. Epub 2021 Sep 16. Arch Biochem Biophys. 2021. PMID: 34506757
-
METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications.Theranostics. 2020 Jul 11;10(20):8939-8956. doi: 10.7150/thno.45178. eCollection 2020. Theranostics. 2020. PMID: 32802173 Free PMC article.
-
METTL14-mediated lncRNA XIST silencing alleviates GDM progression by facilitating trophoblast cell proliferation and migration via the miR-497-5p/FOXO1 axis.J Biochem Mol Toxicol. 2024 Jan;38(1):e23621. doi: 10.1002/jbt.23621. J Biochem Mol Toxicol. 2024. PMID: 38229320
-
Forkhead Box Protein O1: Functional Diversity and Post-Translational Modification, a New Therapeutic Target?Drug Des Devel Ther. 2021 May 3;15:1851-1860. doi: 10.2147/DDDT.S305016. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33976536 Free PMC article. Review.
-
SMC4, a novel tumor prognostic marker and potential tumor therapeutic target.Front Oncol. 2023 Mar 17;13:1117642. doi: 10.3389/fonc.2023.1117642. eCollection 2023. Front Oncol. 2023. PMID: 37007153 Free PMC article. Review.
Cited by
-
N6-Methyladenosine Modification on the Function of Female Reproductive Development and Related Diseases.Immun Inflamm Dis. 2024 Dec;12(12):e70089. doi: 10.1002/iid3.70089. Immun Inflamm Dis. 2024. PMID: 39660878 Free PMC article. Review.
-
Anticancer effects of Erzhimaoling decoction in high-grade serous ovarian cancer in vitro and in vivo.Eur J Med Res. 2024 Aug 5;29(1):405. doi: 10.1186/s40001-024-01968-4. Eur J Med Res. 2024. PMID: 39103890 Free PMC article.
-
Investigation of the mechanism by which FOXJ2 inhibits proliferation, metastasis and cell cycle progression of ovarian cancer cells through the PI3K/AKT signaling pathway.Eur J Med Res. 2025 Mar 4;30(1):152. doi: 10.1186/s40001-025-02270-7. Eur J Med Res. 2025. PMID: 40038842 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

