Pseudo-value regression trees
- PMID: 38403840
- PMCID: PMC11297840
- DOI: 10.1007/s10985-024-09618-x
Pseudo-value regression trees
Abstract
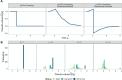
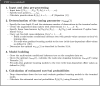
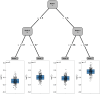
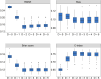
This paper presents a semi-parametric modeling technique for estimating the survival function from a set of right-censored time-to-event data. Our method, named pseudo-value regression trees (PRT), is based on the pseudo-value regression framework, modeling individual-specific survival probabilities by computing pseudo-values and relating them to a set of covariates. The standard approach to pseudo-value regression is to fit a main-effects model using generalized estimating equations (GEE). PRT extend this approach by building a multivariate regression tree with pseudo-value outcome and by successively fitting a set of regularized additive models to the data in the nodes of the tree. Due to the combination of tree learning and additive modeling, PRT are able to perform variable selection and to identify relevant interactions between the covariates, thereby addressing several limitations of the standard GEE approach. In addition, PRT include time-dependent effects in the node-wise models. Interpretability of the PRT fits is ensured by controlling the tree depth. Based on the results of two simulation studies, we investigate the properties of the PRT method and compare it to several alternative modeling techniques. Furthermore, we illustrate PRT by analyzing survival in 3,652 patients enrolled for a randomized study on primary invasive breast cancer.
Keywords: Gradient boosting; Interactions; Model trees; Pseudo-values; Survival probabilities.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Andersen PK, Klein JP, Rosthøj S (2003) Generalised linear models for correlated pseudo-observations, with applications to multi-state models. Biometrika 90:15–27 10.1093/biomet/90.1.15 - DOI
-
- Breiman L, Friedman J, Stone CJ, Olshen RA (1984) Classification and regression trees. Taylor & Francis, New York
MeSH terms
LinkOut - more resources
Full Text Sources

