Proteome analysis develops novel plasma proteins classifier in predicting the mortality of COVID-19
- PMID: 38403992
- PMCID: PMC11216943
- DOI: 10.1111/cpr.13617
Proteome analysis develops novel plasma proteins classifier in predicting the mortality of COVID-19
Abstract
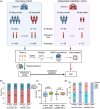
COVID-19 has been a global concern for 3 years, however, consecutive plasma protein changes in the disease course are currently unclear. Setting the mortality within 28 days of admission as the main clinical outcome, plasma samples were collected from patients in discovery and independent validation groups at different time points during the disease course. The whole patients were divided into death and survival groups according to their clinical outcomes. Proteomics and pathway/network analyses were used to find the differentially expressed proteins and pathways. Then, we used machine learning to develop a protein classifier which can predict the clinical outcomes of the patients with COVID-19 and help identify the high-risk patients. Finally, a classifier including C-reactive protein, extracellular matrix protein 1, insulin-like growth factor-binding protein complex acid labile subunit, E3 ubiquitin-protein ligase HECW1 and phosphatidylcholine-sterol acyltransferase was determined. The prediction value of the model was verified with an independent patient cohort. This novel model can realize early prediction of 28-day mortality of patients with COVID-19, with the area under curve 0.88 in discovery group and 0.80 in validation group, superior to 4C mortality and E-CURB65 scores. In total, this work revealed a potential protein classifier which can assist in predicting the outcomes of COVID-19 patients and providing new diagnostic directions.
© 2024 The Authors. Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Machine Learning to Predict Mortality and Critical Events in a Cohort of Patients With COVID-19 in New York City: Model Development and Validation.J Med Internet Res. 2020 Nov 6;22(11):e24018. doi: 10.2196/24018. J Med Internet Res. 2020. PMID: 33027032 Free PMC article.
-
Prognostic accuracy of MALDI-TOF mass spectrometric analysis of plasma in COVID-19.Life Sci Alliance. 2021 Jun 24;4(8):e202000946. doi: 10.26508/lsa.202000946. Print 2021 Aug. Life Sci Alliance. 2021. PMID: 34168074 Free PMC article.
-
A Machine Learning Approach for Mortality Prediction in COVID-19 Pneumonia: Development and Evaluation of the Piacenza Score.J Med Internet Res. 2021 May 31;23(5):e29058. doi: 10.2196/29058. J Med Internet Res. 2021. PMID: 33999838 Free PMC article.
-
Blood proteomics of COVID-19 infection: An update.Clin Chim Acta. 2024 Aug 15;562:119881. doi: 10.1016/j.cca.2024.119881. Epub 2024 Jul 19. Clin Chim Acta. 2024. PMID: 39033952 Review.
-
Benchmarking of Machine Learning classifiers on plasma proteomic for COVID-19 severity prediction through interpretable artificial intelligence.Artif Intell Med. 2023 Mar;137:102490. doi: 10.1016/j.artmed.2023.102490. Epub 2023 Jan 18. Artif Intell Med. 2023. PMID: 36868685 Free PMC article. Review.
Cited by
-
Utility of Protein Markers in COVID-19 Patients.Int J Mol Sci. 2025 Jan 14;26(2):653. doi: 10.3390/ijms26020653. Int J Mol Sci. 2025. PMID: 39859366 Free PMC article. Review.
-
Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis.Viruses. 2025 Jun 23;17(7):882. doi: 10.3390/v17070882. Viruses. 2025. PMID: 40733500 Free PMC article. Review.
References
-
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239‐1242. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

