Capturing chromosome conformation in Crenarchaea
- PMID: 38404013
- PMCID: PMC11344861
- DOI: 10.1111/mmi.15245
Capturing chromosome conformation in Crenarchaea
Abstract
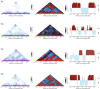
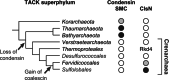
While there is a considerable body of knowledge regarding the molecular and structural biology and biochemistry of archaeal information processing machineries, far less is known about the nature of the substrate for these machineries-the archaeal nucleoid. In this article, we will describe recent advances in our understanding of the three-dimensional organization of the chromosomes of model organisms in the crenarchaeal phylum.
Keywords: Sulfolobus; Archaea; SMC; condensin; evolution; genome.
© 2024 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

