Comprehensive integration of single-cell RNA and transcriptome RNA sequencing to establish a pyroptosis-related signature for improving prognostic prediction of gastric cancer
- PMID: 38404710
- PMCID: PMC10884435
- DOI: 10.1016/j.csbj.2024.02.002
Comprehensive integration of single-cell RNA and transcriptome RNA sequencing to establish a pyroptosis-related signature for improving prognostic prediction of gastric cancer
Abstract
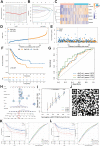
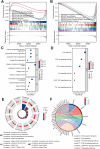
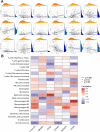
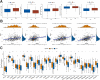
Cell pyroptosis, a Gasdermin-dependent programmed cell death characterized by inflammasome, plays a complex and dynamic role in Gastric cancer (GC), a serious threat to human health. Therefore, the value of pyroptosis-related genes (PRGs) as prognostic biomarkers and therapeutic indicators for patients needs to be exploited in GC. This study integrates single-cell RNA sequencing (scRNA-seq) dataset GSE183904 with GC transcriptome data from the TCGA database, focusing on the expression and distribution of PRGs in GC at the single-cell level. The prognostic signature of PRGs was established by using Cox and LASSO analyses. The differences in long-term prognosis, immune infiltration, mutation profile, CD274 and response to chemotherapeutic drugs between the two groups were analyzed and evaluated. A tissue array was used to verify the expression of six PRGs, CD274, CD163 and FoxP3. C12orf75, VCAN, RGS2, MKNK2, SOCS3 and TNFAIP2 were successfully screened out to establish a signature to potently predict the survival time of GC patients. A webserver (https://pumc.shinyapps.io/GastricCancer/) for prognostic prediction in GC patients was developed based on this signature. High-risk score patients typically had worse prognoses, resistance to classical chemotherapy, and a more immunosuppressive tumor microenvironment. VCAN, TNFAIP2 and SOCS3 were greatly elevated in the GC while RGS2 and MKNK2 were decreased in the tumor samples. Further, VCAN was positively related to the infiltrations of Tregs and M2 TAMs in GC TME and the CD274 in tumor cells. In summary, a potent pyroptosis-related signature was established to accurately forecast the survival time and treatment responsiveness of GC patients.
Keywords: Cell pyroptosis; Gastric cancer; Immune infiltration; Prognostic signature; Single-cell RNA sequencing.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Integrated analysis of disulfidptosis-related immune genes signature to boost the efficacy of prognostic prediction in gastric cancer.Cancer Cell Int. 2024 Mar 25;24(1):112. doi: 10.1186/s12935-024-03294-5. Cancer Cell Int. 2024. PMID: 38528532 Free PMC article.
-
Pyroptosis-Related Gene Signature Predicts Prognosis and Indicates Immune Microenvironment Infiltration in Glioma.Front Cell Dev Biol. 2022 Apr 25;10:862493. doi: 10.3389/fcell.2022.862493. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35547808 Free PMC article.
-
Development of a CD8+ T cell associated signature for predicting the prognosis and immunological characteristics of gastric cancer by integrating single-cell and bulk RNA-sequencing.Sci Rep. 2024 Feb 24;14(1):4524. doi: 10.1038/s41598-024-54273-9. Sci Rep. 2024. PMID: 38402299 Free PMC article.
-
Systematic transcriptome profiling of pyroptosis related signature for predicting prognosis and immune landscape in lower grade glioma.BMC Cancer. 2022 Aug 13;22(1):885. doi: 10.1186/s12885-022-09982-7. BMC Cancer. 2022. PMID: 35964070 Free PMC article.
-
Comprehensive analysis of cuproptosis-related immune biomarker signature to enhance prognostic accuracy in gastric cancer.Aging (Albany NY). 2023 Apr 7;15(7):2772-2796. doi: 10.18632/aging.204646. Epub 2023 Apr 7. Aging (Albany NY). 2023. PMID: 37036489 Free PMC article.
Cited by
-
A signature based on efferocytosis-related genes for the evaluation of prognosis and the tumour microenvironment in gastric cancer.Sci Rep. 2025 Apr 24;15(1):14226. doi: 10.1038/s41598-025-99133-2. Sci Rep. 2025. PMID: 40275059 Free PMC article.
-
Comprehensive analysis of single-cell and bulk RNA sequencing data unveils antigen-presenting and processing fibroblasts and establishes a predictive model in gastric cancer.Cancer Cell Int. 2025 Jun 21;25(1):225. doi: 10.1186/s12935-025-03878-9. Cancer Cell Int. 2025. PMID: 40544264 Free PMC article.
-
TNFAIP2 promotes NF-κB signaling mediate lymph node metastasis of oral squamous cell carcinoma by protecting IKKβ from ubiquitin proteasome degradation.Cell Commun Signal. 2025 Feb 13;23(1):83. doi: 10.1186/s12964-025-02077-3. Cell Commun Signal. 2025. PMID: 39948570 Free PMC article.
-
Establishing a Novel Pyroptosis-related Gene Signature and Predicting Chemical Drugs for Papillary Thyroid Cancer.Curr Pharm Biotechnol. 2025;26(8):1232-1244. doi: 10.2174/0113892010325685241029113633. Curr Pharm Biotechnol. 2025. PMID: 39492777
-
Pyroptosis-Related Gene Signatures Enable Robustly Diagnosis, Prognosis and Immune Responses Prediction in Uterine Corpus Endometrial Carcinoma.J Cancer. 2025 May 8;16(8):2516-2536. doi: 10.7150/jca.104826. eCollection 2025. J Cancer. 2025. PMID: 40535818 Free PMC article.
References
-
- Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Johnston F.M., Beckman M. Updates on management of gastric cancer. Curr Oncol Rep. 2019;21(8):67. - PubMed
-
- Janjigian Y.Y., Shitara K., Moehler M., et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet (Lond, Engl) 2021;398(10294):27–40. - PMC - PubMed
-
- Bang Y.J., Van Cutsem E., Fuchs C.S., et al. KEYNOTE-585: phase III study of perioperative chemotherapy with or without pembrolizumab for gastric cancer. Future Oncol (Lond, Engl) 2019;15(9):943–952. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
