Perspectives on Computational Enzyme Modeling: From Mechanisms to Design and Drug Development
- PMID: 38405524
- PMCID: PMC10883025
- DOI: 10.1021/acsomega.3c09084
Perspectives on Computational Enzyme Modeling: From Mechanisms to Design and Drug Development
Abstract
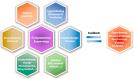
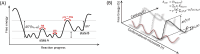
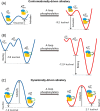
Understanding enzyme mechanisms is essential for unraveling the complex molecular machinery of life. In this review, we survey the field of computational enzymology, highlighting key principles governing enzyme mechanisms and discussing ongoing challenges and promising advances. Over the years, computer simulations have become indispensable in the study of enzyme mechanisms, with the integration of experimental and computational exploration now established as a holistic approach to gain deep insights into enzymatic catalysis. Numerous studies have demonstrated the power of computer simulations in characterizing reaction pathways, transition states, substrate selectivity, product distribution, and dynamic conformational changes for various enzymes. Nevertheless, significant challenges remain in investigating the mechanisms of complex multistep reactions, large-scale conformational changes, and allosteric regulation. Beyond mechanistic studies, computational enzyme modeling has emerged as an essential tool for computer-aided enzyme design and the rational discovery of covalent drugs for targeted therapies. Overall, enzyme design/engineering and covalent drug development can greatly benefit from our understanding of the detailed mechanisms of enzymes, such as protein dynamics, entropy contributions, and allostery, as revealed by computational studies. Such a convergence of different research approaches is expected to continue, creating synergies in enzyme research. This review, by outlining the ever-expanding field of enzyme research, aims to provide guidance for future research directions and facilitate new developments in this important and evolving field.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Modeling Catalysis in Allosteric Enzymes: Capturing Conformational Consequences.Top Catal. 2022 Feb;65(1-4):165-186. doi: 10.1007/s11244-021-01521-1. Epub 2021 Nov 9. Top Catal. 2022. PMID: 36304771 Free PMC article.
-
Advances in the Directed Evolution of Computer-aided Enzymes.Curr Top Med Chem. 2025 May 29. doi: 10.2174/0115680266377310250526045137. Online ahead of print. Curr Top Med Chem. 2025. PMID: 40454510
-
Hybrid schemes based on quantum mechanics/molecular mechanics simulations goals to success, problems, and perspectives.Adv Protein Chem Struct Biol. 2011;85:81-142. doi: 10.1016/B978-0-12-386485-7.00003-X. Adv Protein Chem Struct Biol. 2011. PMID: 21920322 Review.
-
Exploring and Learning the Universe of Protein Allostery Using Artificial Intelligence Augmented Biophysical and Computational Approaches.J Chem Inf Model. 2023 Mar 13;63(5):1413-1428. doi: 10.1021/acs.jcim.2c01634. Epub 2023 Feb 24. J Chem Inf Model. 2023. PMID: 36827465 Free PMC article. Review.
-
Understanding G Protein-Coupled Receptor Allostery via Molecular Dynamics Simulations: Implications for Drug Discovery.Methods Mol Biol. 2018;1762:455-472. doi: 10.1007/978-1-4939-7756-7_23. Methods Mol Biol. 2018. PMID: 29594786
Cited by
-
Fermi calculations enable quick downselection of target genes and process optimization in photosynthetic systems.Plant Physiol. 2025 Jul 3;198(3):kiaf103. doi: 10.1093/plphys/kiaf103. Plant Physiol. 2025. PMID: 40112183 Free PMC article. Review.
-
Magnesium induced structural reorganization in the active site of adenylate kinase.Sci Adv. 2024 Aug 9;10(32):eado5504. doi: 10.1126/sciadv.ado5504. Epub 2024 Aug 9. Sci Adv. 2024. PMID: 39121211 Free PMC article.
-
Computation of Protein-Ligand Binding Free Energies with a Quantum Mechanics-Based Mining Minima Algorithm.J Chem Theory Comput. 2025 Apr 22;21(8):4236-4265. doi: 10.1021/acs.jctc.4c01707. Epub 2025 Apr 9. J Chem Theory Comput. 2025. PMID: 40202178 Free PMC article.
-
Microbial lipases: advances in production, purification, biochemical characterization, and multifaceted applications in industry and medicine.Microb Cell Fact. 2025 Feb 12;24(1):40. doi: 10.1186/s12934-025-02664-6. Microb Cell Fact. 2025. PMID: 39939876 Free PMC article. Review.
-
CHARMM at 45: Enhancements in Accessibility, Functionality, and Speed.J Phys Chem B. 2024 Oct 17;128(41):9976-10042. doi: 10.1021/acs.jpcb.4c04100. Epub 2024 Sep 20. J Phys Chem B. 2024. PMID: 39303207 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
