Noninvasive total counting of cultured cells using a home-use scanner with a pattern sheet
- PMID: 38405610
- PMCID: PMC10884908
- DOI: 10.1016/j.isci.2024.109170
Noninvasive total counting of cultured cells using a home-use scanner with a pattern sheet
Abstract
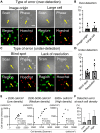
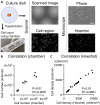
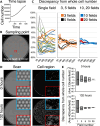
The inherent variability in cell culture techniques hinders their reproducibility. To address this issue, we introduce a comprehensive cell observation device. This new approach enhances the features of existing home-use scanners by implementing a pattern sheet. Compared with fluorescent staining, our method over- or underestimated the cell count by a mere 5%. The proposed technique showcased a strong correlation with conventional methodologies, displaying R2 values of 0.91 and 0.99 compared with the standard chamber and fluorescence methods, respectively. Simulations of microscopic observations indicated the potential to estimate accurately the total cell count using just 20 fields of view. Our proposed cell-counting device offers a straightforward, noninvasive means of measuring the number of cultured cells. By harnessing the power of deep learning, this device ensures data integrity, thereby making it an attractive option for future cell culture research.
Keywords: Biotechnology; Machine learning; Optical imaging.
© 2024 The Author(s).
Conflict of interest statement
A patent application for this technology has been submitted in Japan.
Figures





Similar articles
-
Estimation of absolute number of alveolar epithelial type 2 cells in mouse lungs: a comparison between stereology and flow cytometry.J Microsc. 2019 Jul;275(1):36-50. doi: 10.1111/jmi.12800. Epub 2019 May 6. J Microsc. 2019. PMID: 31020994
-
Automated White Blood Cell Counting in Nailfold Capillary Using Deep Learning Segmentation and Video Stabilization.Sensors (Basel). 2020 Dec 11;20(24):7101. doi: 10.3390/s20247101. Sensors (Basel). 2020. PMID: 33322435 Free PMC article.
-
Validation of analytical methods in GMP: the disposable Fast Read 102® device, an alternative practical approach for cell counting.J Transl Med. 2012 May 31;10:112. doi: 10.1186/1479-5876-10-112. J Transl Med. 2012. PMID: 22650233 Free PMC article.
-
[Aiming for zero blindness].Nippon Ganka Gakkai Zasshi. 2015 Mar;119(3):168-93; discussion 194. Nippon Ganka Gakkai Zasshi. 2015. PMID: 25854109 Review. Japanese.
-
A multi-view time-domain non-contact diffuse optical tomography scanner with dual wavelength detection for intrinsic and fluorescence small animal imaging.Rev Sci Instrum. 2012 Jun;83(6):063703. doi: 10.1063/1.4726016. Rev Sci Instrum. 2012. PMID: 22755630 Review.
References
LinkOut - more resources
Full Text Sources

