This is a preprint.
Scalable nonparametric clustering with unified marker gene selection for single-cell RNA-seq data
- PMID: 38405697
- PMCID: PMC10888887
- DOI: 10.1101/2024.02.11.579839
Scalable nonparametric clustering with unified marker gene selection for single-cell RNA-seq data
Abstract
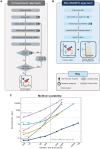
Clustering is commonly used in single-cell RNA-sequencing (scRNA-seq) pipelines to characterize cellular heterogeneity. However, current methods face two main limitations. First, they require user-specified heuristics which add time and complexity to bioinformatic workflows; second, they rely on post-selective differential expression analyses to identify marker genes driving cluster differences, which has been shown to be subject to inflated false discovery rates. We address these challenges by introducing nonparametric clustering of single-cell populations (NCLUSION): an infinite mixture model that leverages Bayesian sparse priors to identify marker genes while simultaneously performing clustering on single-cell expression data. NCLUSION uses a scalable variational inference algorithm to perform these analyses on datasets with up to millions of cells. Through simulations and analyses of publicly available scRNA-seq studies, we demonstrate that NCLUSION (i) matches the performance of other state-of-the-art clustering techniques with significantly reduced runtime and (ii) provides statistically robust and biologically relevant transcriptomic signatures for each of the clusters it identifies. Overall, NCLUSION represents a reliable hypothesis-generating tool for understanding patterns of expression variation present in single-cell populations.
Conflict of interest statement
SR holds equity in Amgen. PSW reports compensation for consulting/speaking from Engine Ventures and AbbVie unrelated to this work. AKS reports compensation for consulting and/or scientific advisory board membership from Honeycomb Biotechnologies, Cellarity, Ochre Bio, Relation Therapeutics, Fog Pharma, Bio-Rad Laboratories, IntrECate Biotherapeutics, Passkey Therapeutics and Dahlia Biosciences unrelated to this work. SR and PSW receive research funding from Microsoft. MH, NF, APA, and LC are employees of Microsoft and own equity in Microsoft. All other authors have declared that no competing interests exist.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
