This is a preprint.
Host-Microbe Multiomic Profiling Reveals Age-Dependent COVID-19 Immunopathology
- PMID: 38405760
- PMCID: PMC10888993
- DOI: 10.1101/2024.02.11.24301704
Host-Microbe Multiomic Profiling Reveals Age-Dependent COVID-19 Immunopathology
Update in
-
Host-microbe multiomic profiling reveals age-dependent immune dysregulation associated with COVID-19 immunopathology.Sci Transl Med. 2024 Apr 17;16(743):eadj5154. doi: 10.1126/scitranslmed.adj5154. Epub 2024 Apr 17. Sci Transl Med. 2024. PMID: 38630846 Free PMC article.
Abstract
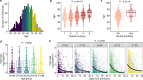
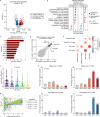
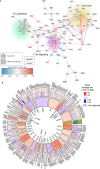
Age is a major risk factor for severe coronavirus disease-2019 (COVID-19), yet the mechanisms responsible for this relationship have remained incompletely understood. To address this, we evaluated the impact of aging on host and viral dynamics in a prospective, multicenter cohort of 1,031 patients hospitalized for COVID-19, ranging from 18 to 96 years of age. We performed blood transcriptomics and nasal metatranscriptomics, and measured peripheral blood immune cell populations, inflammatory protein expression, anti-SARS-CoV-2 antibodies, and anti-interferon (IFN) autoantibodies. We found that older age correlated with an increased SARS-CoV-2 viral load at the time of admission, and with delayed viral clearance over 28 days. This contributed to an age-dependent increase in type I IFN gene expression in both the respiratory tract and blood. We also observed age-dependent transcriptional increases in peripheral blood IFN-γ, neutrophil degranulation, and Toll like receptor (TLR) signaling pathways, and decreases in T cell receptor (TCR) and B cell receptor signaling pathways. Over time, older adults exhibited a remarkably sustained induction of proinflammatory genes (e.g., CXCL6) and serum chemokines (e.g., CXCL9) compared to younger individuals, highlighting a striking age-dependent impairment in inflammation resolution. Augmented inflammatory signaling also involved the upper airway, where aging was associated with upregulation of TLR, IL17, type I IFN and IL1 pathways, and downregulation TCR and PD-1 signaling pathways. Metatranscriptomics revealed that the oldest adults exhibited disproportionate reactivation of herpes simplex virus and cytomegalovirus in the upper airway following hospitalization. Mass cytometry demonstrated that aging correlated with reduced naïve T and B cell populations, and increased monocytes and exhausted natural killer cells. Transcriptional and protein biomarkers of disease severity markedly differed with age, with the oldest adults exhibiting greater expression of TLR and inflammasome signaling genes, as well as proinflammatory proteins (e.g., IL6, CXCL8), in severe COVID-19 compared to mild/moderate disease. Anti-IFN autoantibody prevalence correlated with both age and disease severity. Taken together, this work profiles both host and microbe in the blood and airway to provide fresh insights into aging-related immune changes in a large cohort of vaccine-naïve COVID-19 patients. We observed age-dependent immune dysregulation at the transcriptional, protein and cellular levels, manifesting in an imbalance of inflammatory responses over the course of hospitalization, and suggesting potential new therapeutic targets.
Figures





References
-
- O’Driscoll M. et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 590, 140–145 (2021). - PubMed
-
- United States Centers for Disease Control and Prevention. Risk for COVID-19 Infection, Hospitalization, and Death By Age Group. Risk for COVID-19 Infection, Hospitalization, and Death By Age Group https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-disc... (2022).
Publication types
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- U19 AI167891/AI/NIAID NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- R01 AI132774/AI/NIAID NIH HHS/United States
- R01 AI122220/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
