This is a preprint.
The budding yeast Fkh1 Forkhead associated (FHA) domain promoted a G1-chromatin state and the activity of chromosomal DNA replication origins
- PMID: 38405780
- PMCID: PMC10889021
- DOI: 10.1101/2024.02.16.580712
The budding yeast Fkh1 Forkhead associated (FHA) domain promoted a G1-chromatin state and the activity of chromosomal DNA replication origins
Update in
-
The budding yeast Fkh1 Forkhead associated (FHA) domain promotes a G1-chromatin state and the activity of chromosomal DNA replication origins.PLoS Genet. 2024 Aug 5;20(8):e1011366. doi: 10.1371/journal.pgen.1011366. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39102423 Free PMC article.
Abstract
In Saccharomyces cerevisiae, the forkhead (Fkh) transcription factor Fkh1 (forkhead homolog) enhances the activity of many DNA replication origins that act in early S-phase (early origins). Current models posit that Fkh1 acts directly to promote these origins' activity by binding to origin-adjacent Fkh1 binding sites (FKH sites). However, the post-DNA binding functions that Fkh1 uses to promote early origin activity are poorly understood. Fkh1 contains a conserved FHA (forkhead associated) domain, a protein-binding module with specificity for phosphothreonine (pT)-containing partner proteins. At a small subset of yeast origins, the Fkh1-FHA domain enhances the ORC (origin recognition complex)-origin binding step, the G1-phase event that initiates the origin cycle. However, the importance of the Fkh1-FHA domain to either chromosomal replication or ORC-origin interactions at genome scale is unclear. Here, S-phase SortSeq experiments were used to compare genome replication in proliferating FKH1 and fkh1-R80A mutant cells. The Fkh1-FHA domain promoted the activity of 100 origins that act in early to mid- S-phase, including the majority of centromere-associated origins, while simultaneously inhibiting 100 late origins. Thus, in the absence of a functional Fkh1-FHA domain, the temporal landscape of the yeast genome was flattened. Origins are associated with a positioned nucleosome array that frames a nucleosome depleted region (NDR) over the origin, and ORC-origin binding is necessary but not sufficient for this chromatin organization. To ask whether the Fkh1-FHA domain had an impact on this chromatin architecture at origins, ORC ChIPSeq data generated from proliferating cells and MNaseSeq data generated from G1-arrested and proliferating cell populations were assessed. Origin groups that were differentially regulated by the Fkh1-FHA domain were characterized by distinct effects of this domain on ORC-origin binding and G1-phase chromatin. Thus, the Fkh1-FHA domain controlled the distinct chromatin architecture at early origins in G1-phase and regulated origin activity in S-phase.
Conflict of interest statement
Conflict of interest statement None declared.
Figures


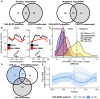



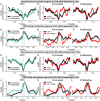
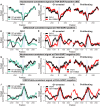
Similar articles
-
The budding yeast Fkh1 Forkhead associated (FHA) domain promotes a G1-chromatin state and the activity of chromosomal DNA replication origins.PLoS Genet. 2024 Aug 5;20(8):e1011366. doi: 10.1371/journal.pgen.1011366. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39102423 Free PMC article.
-
The Fkh1 Forkhead associated domain promotes ORC binding to a subset of DNA replication origins in budding yeast.Nucleic Acids Res. 2021 Oct 11;49(18):10207-10220. doi: 10.1093/nar/gkab450. Nucleic Acids Res. 2021. PMID: 34095951 Free PMC article.
-
Binding of the Fkh1 Forkhead Associated Domain to a Phosphopeptide within the Mph1 DNA Helicase Regulates Mating-Type Switching in Budding Yeast.PLoS Genet. 2016 Jun 3;12(6):e1006094. doi: 10.1371/journal.pgen.1006094. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27257873 Free PMC article.
-
Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review.
-
Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle.J Cell Sci Suppl. 1995;19:67-72. doi: 10.1242/jcs.1995.supplement_19.9. J Cell Sci Suppl. 1995. PMID: 8655649 Review.
References
-
- Costa A, Diffley JFX. The Initiation of Eukaryotic DNA Replication. Annu Rev Biochem. 2022;91: 107–131. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
