This is a preprint.
Functional Characterisation of the ATOH1 Molecular Subtype Indicates a Pro-Metastatic Role in Small Cell Lung Cancer
- PMID: 38405859
- PMCID: PMC10888785
- DOI: 10.1101/2024.02.16.580247
Functional Characterisation of the ATOH1 Molecular Subtype Indicates a Pro-Metastatic Role in Small Cell Lung Cancer
Update in
-
Functional characterization of the ATOH1 molecular subtype indicates a pro-metastatic role in small cell lung cancer.Cell Rep. 2025 May 27;44(5):115603. doi: 10.1016/j.celrep.2025.115603. Epub 2025 Apr 29. Cell Rep. 2025. PMID: 40305287 Free PMC article.
Abstract
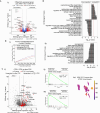
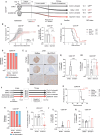
Molecular subtypes of Small Cell Lung Cancer (SCLC) have been described based on differential expression of transcription factors (TFs) ASCL1, NEUROD1, POU2F3 and immune-related genes. We previously reported an additional subtype based on expression of the neurogenic TF ATOH1 within our SCLC Circulating tumour cell-Derived eXplant (CDX) model biobank. Here we show that ATOH1 protein was detected in 7/81 preclinical models and 16/102 clinical samples of SCLC. In CDX models, ATOH1 directly regulated neurogenesis and differentiation programs consistent with roles in normal tissues. In ex vivo cultures of ATOH1-positive CDX, ATOH1 was required for cell survival. In vivo, ATOH1 depletion slowed tumour growth and suppressed liver metastasis. Our data validate ATOH1 as a bona fide oncogenic driver of SCLC with tumour cell survival and pro-metastatic functions. Further investigation to explore ATOH1 driven vulnerabilities for targeted treatment with predictive biomarkers is warranted.
Figures




References
-
- Forman D. Cancer incidence in five continents: volume X. (International Agency for Research on Cancer, 2014).
-
- American Cancer Society Cancer Facts & Figures 2022. (2022).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
