This is a preprint.
Bending DNA increases its helical repeat
- PMID: 38405957
- PMCID: PMC10888926
- DOI: 10.1101/2024.02.14.579968
Bending DNA increases its helical repeat
Abstract
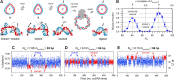
In all biological systems, DNA is under high mechanical stress from bending and twisting. For example, DNA is tightly bent in nucleosome complexes, virus capsids, bacterial chromosomes, or complexes with transcription factors that regulate gene expression. A structurally and mechanically accurate model of DNA is therefore necessary to understand some of the most fundamental molecular mechanisms in biology including DNA packaging, replication, transcription and gene regulation. An iconic feature of DNA is its double helical nature with an average repeat of ~10.45 base pairs per turn, which is commonly believed to be independent of curvature. We developed a ligation assay on nicked DNA circles of variable curvature that reveals a strong unwinding of DNA to over 11 bp/turn for radii around 3-4 nm. Our work constitutes a major modification of the standard mechanical model of DNA and requires reassessing the molecular mechanisms and energetics of all processes involving tightly bent DNA.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests.
Figures


References
-
- Basu A.; Bobrovnikov D. G.; Ha T. DNA Mechanics and Its Biological Impact. J. Mol. Biol. 2021, 433 (6), 166861. - PubMed
-
- Shimada J.; Yamakawa H. Ring-Closure Probabilities for Twisted Wormlike Chains. Application to DNA. Macromolecules 1984, 17 (4), 689–698.
-
- Shore D.; Baldwin R. L. Energetics of DNA Twisting: II. Topoisomer Analysis. J. Mol. Biol. 1983, 170 (4), 983–1007. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
