This is a preprint.
Structural and biochemical characterization of the mitomycin C repair exonuclease MrfB
- PMID: 38405983
- PMCID: PMC10889028
- DOI: 10.1101/2024.02.15.580553
Structural and biochemical characterization of the mitomycin C repair exonuclease MrfB
Update in
-
Structural and biochemical characterization of the mitomycin C repair exonuclease MrfB.Nucleic Acids Res. 2024 Jun 24;52(11):6347-6359. doi: 10.1093/nar/gkae308. Nucleic Acids Res. 2024. PMID: 38661211 Free PMC article.
Abstract
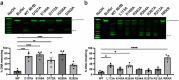
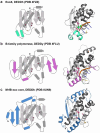
Mitomycin C (MMC) repair factor A (mrfA) and factor B (mrfB), encode a conserved helicase and exonuclease that repair DNA damage in the soil-dwelling bacterium Bacillus subtilis. Here we have focused on the characterization of MrfB, a DEDDh exonuclease in the DnaQ superfamily. We solved the structure of the exonuclease core of MrfB to a resolution of 2.1 Å, in what appears to be an inactive state. In this conformation, a predicted α-helix containing the catalytic DEDDh residue Asp172 adopts a random coil, which moves Asp172 away from the active site and results in the occupancy of only one of the two catalytic Mg2+ ions. We propose that MrfB resides in this inactive state until it interacts with DNA to become activated. By comparing our structure to an AlphaFold prediction as well as other DnaQ-family structures, we located residues hypothesized to be important for exonuclease function. Using exonuclease assays we show that MrfB is a Mg2+-dependent 3'-5' DNA exonuclease. We show that Leu113 aids in coordinating the 3' end of the DNA substrate, and that a basic loop is important for substrate binding. This work provides insight into the function of a recently discovered bacterial exonuclease important for the repair of MMC-induced DNA adducts.
Figures





Similar articles
-
Structural and biochemical characterization of the mitomycin C repair exonuclease MrfB.Nucleic Acids Res. 2024 Jun 24;52(11):6347-6359. doi: 10.1093/nar/gkae308. Nucleic Acids Res. 2024. PMID: 38661211 Free PMC article.
-
A bacterial DNA repair pathway specific to a natural antibiotic.Mol Microbiol. 2019 Feb;111(2):338-353. doi: 10.1111/mmi.14158. Epub 2018 Nov 28. Mol Microbiol. 2019. PMID: 30379365 Free PMC article.
-
Exonuclease X of Escherichia coli. A novel 3'-5' DNase and Dnaq superfamily member involved in DNA repair.J Biol Chem. 1999 Oct 15;274(42):30094-100. doi: 10.1074/jbc.274.42.30094. J Biol Chem. 1999. PMID: 10514496
-
DNA stabilization at the Bacillus subtilis PolX core--a binding model to coordinate polymerase, AP-endonuclease and 3'-5' exonuclease activities.Nucleic Acids Res. 2012 Oct;40(19):9750-62. doi: 10.1093/nar/gks702. Epub 2012 Jul 25. Nucleic Acids Res. 2012. PMID: 22844091 Free PMC article.
-
Understanding APE1 cellular functions by the structural preference of exonuclease activities.Comput Struct Biotechnol J. 2021 Jun 24;19:3682-3691. doi: 10.1016/j.csbj.2021.06.036. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285771 Free PMC article. Review.
References
-
- Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A. and Ellenberger T. (2006) DNA Repair and Mutagenesis: Second Edition. American Society for Microbiology, Washington, DC.
-
- Procopio R.E., Silva I.R., Martins M.K., Azevedo J.L. and Araujo J.M. (2012) Antibiotics produced by Streptomyces. Braz J Infect Dis, 16, 466–471. - PubMed
-
- Hata T., Sano Y., Sugawara R., Matsumae A., Kanamori K., Shima T. and Hoshi T. (1956) Mitomycin, a new antibiotic from Streptomyces. I. The Journal of Antibiotics, Series A, 9, 141–146. - PubMed
-
- Tomasz M. (1995) Mitomycin C: small, fast and deadly (but very selective). Chemistry & biology, 2, 575–579. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
