This is a preprint.
Complementary Cytoskeletal Feedback Loops Control Signal Transduction Excitability and Cell Polarity
- PMID: 38405988
- PMCID: PMC10888828
- DOI: 10.1101/2024.02.13.580131
Complementary Cytoskeletal Feedback Loops Control Signal Transduction Excitability and Cell Polarity
Update in
-
Complementary cytoskeletal feedback loops control signal transduction excitability and cell polarity.Nat Commun. 2025 Aug 12;16(1):7482. doi: 10.1038/s41467-025-62799-3. Nat Commun. 2025. PMID: 40796751 Free PMC article.
Abstract
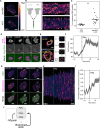
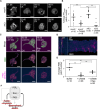
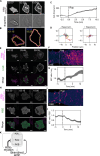
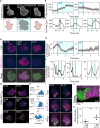
To move through complex environments, cells must constantly integrate chemical and mechanical cues. Signaling networks, such as those comprising Ras and PI3K, transmit chemical cues to the cytoskeleton, but the cytoskeleton must also relay mechanical information back to those signaling systems. Using novel synthetic tools to acutely control specific elements of the cytoskeleton in Dictyostelium and neutrophils, we delineate feedback mechanisms that alter the signaling network and promote front- or back-states of the cell membrane and cortex. First, increasing branched actin assembly increases Ras/PI3K activation while reducing polymeric actin levels overall decreases activation. Second, reducing myosin II assembly immediately increases Ras/PI3K activation and sensitivity to chemotactic stimuli. Third, inhibiting branched actin alone increases cortical actin assembly and strongly blocks Ras/PI3K activation. This effect is mitigated by reducing filamentous actin levels and in cells lacking myosin II. Finally, increasing actin crosslinking with a controllable activator of cytoskeletal regulator RacE leads to a large decrease in Ras activation both globally and locally. Curiously, RacE activation can trigger cell spreading and protrusion with no detectable activation of branched actin nucleators. Taken together with legacy data that Ras/PI3K promotes branched actin assembly and myosin II disassembly, our results define front- and back-promoting positive feedback loops. We propose that these loops play a crucial role in establishing cell polarity and mediating signal integration by controlling the excitable state of the signal transduction networks in respective regions of the membrane and cortex. This interplay enables cells to navigate intricate topologies like tissues containing other cells, the extracellular matrix, and fluids.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
