Catalytic divergencies in the mechanism of L-arginine hydroxylating nonheme iron enzymes
- PMID: 38406558
- PMCID: PMC10884159
- DOI: 10.3389/fchem.2024.1365494
Catalytic divergencies in the mechanism of L-arginine hydroxylating nonheme iron enzymes
Abstract
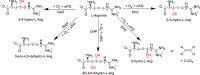
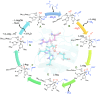
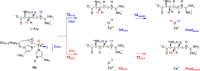
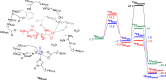
Many enzymes in nature utilize a free arginine (L-Arg) amino acid to initiate the biosynthesis of natural products. Examples include nitric oxide synthases, which generate NO from L-Arg for blood pressure control, and various arginine hydroxylases involved in antibiotic biosynthesis. Among the groups of arginine hydroxylases, several enzymes utilize a nonheme iron(II) active site and let L-Arg react with dioxygen and α-ketoglutarate to perform either C3-hydroxylation, C4-hydroxylation, C5-hydroxylation, or C4-C5-desaturation. How these seemingly similar enzymes can react with high specificity and selectivity to form different products remains unknown. Over the past few years, our groups have investigated the mechanisms of L-Arg-activating nonheme iron dioxygenases, including the viomycin biosynthesis enzyme VioC, the naphthyridinomycin biosynthesis enzyme NapI, and the streptothricin biosynthesis enzyme OrfP, using computational approaches and applied molecular dynamics, quantum mechanics on cluster models, and quantum mechanics/molecular mechanics (QM/MM) approaches. These studies not only highlight the differences in substrate and oxidant binding and positioning but also emphasize on electronic and electrostatic differences in the substrate-binding pockets of the enzymes. In particular, due to charge differences in the active site structures, there are changes in the local electric field and electric dipole moment orientations that either strengthen or weaken specific substrate C-H bonds. The local field effects, therefore, influence and guide reaction selectivity and specificity and give the enzymes their unique reactivity patterns. Computational work using either QM/MM or density functional theory (DFT) on cluster models can provide valuable insights into catalytic reaction mechanisms and produce accurate and reliable data that can be used to engineer proteins and synthetic catalysts to perform novel reaction pathways.
Keywords: QM/MM; cluster models; dioxygenases; enzyme catalysis; inorganic reaction mechanisms; iron enzymes.
Copyright © 2024 Ali and de Visser.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declare that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures






References
-
- Ali H. S., Henchman R. H., Warwicker J., de Visser S. P. (2021a). How do electrostatic perturbations of the protein affect the bifurcation pathways of substrate hydroxylation versus desaturation in the nonheme iron-dependent viomycin biosynthesis enzyme? J. Phys. Chem. A 125, 1720–1737. 10.1021/acs.jpca.1c00141 - DOI - PubMed
-
- Ali H. S., Warwicker J., de Visser S. P. (2023). How does the nonheme iron enzyme NapI react through l-arginine desaturation rather than hydroxylation? A quantum mechanics/molecular mechanics study. ACS Catal. 13, 10705–10721. 10.1021/acscatal.3c02262 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

