Recent advance of microbial mercury methylation in the environment
- PMID: 38407657
- PMCID: PMC10896945
- DOI: 10.1007/s00253-023-12967-6
Recent advance of microbial mercury methylation in the environment
Abstract
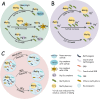
Methylmercury formation is mainly driven by microbial-mediated process. The mechanism of microbial mercury methylation has become a crucial research topic for understanding methylation in the environment. Pioneering studies of microbial mercury methylation are focusing on functional strain isolation, microbial community composition characterization, and mechanism elucidation in various environments. Therefore, the functional genes of microbial mercury methylation, global isolations of Hg methylation strains, and their methylation potential were systematically analyzed, and methylators in typical environments were extensively reviewed. The main drivers (key physicochemical factors and microbiota) of microbial mercury methylation were summarized and discussed. Though significant progress on the mechanism of the Hg microbial methylation has been explored in recent decade, it is still limited in several aspects, including (1) molecular biology techniques for identifying methylators; (2) characterization methods for mercury methylation potential; and (3) complex environmental properties (environmental factors, complex communities, etc.). Accordingly, strategies for studying the Hg microbial methylation mechanism were proposed. These strategies include the following: (1) the development of new molecular biology methods to characterize methylation potential; (2) treating the environment as a micro-ecosystem and studying them from a holistic perspective to clearly understand mercury methylation; (3) a more reasonable and sensitive inhibition test needs to be considered. KEY POINTS: • Global Hg microbial methylation is phylogenetically and functionally discussed. • The main drivers of microbial methylation are compared in various condition. • Future study of Hg microbial methylation is proposed.
Keywords: Driving factors; Functional genes; Functional strains; Mercury methylation; Methylation mechanism; Methylation potential.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Periphyton and Flocculent Materials Are Important Ecological Compartments Supporting Abundant and Diverse Mercury Methylator Assemblages in the Florida Everglades.Appl Environ Microbiol. 2019 Jun 17;85(13):e00156-19. doi: 10.1128/AEM.00156-19. Print 2019 Jul 1. Appl Environ Microbiol. 2019. PMID: 31028023 Free PMC article.
-
Understanding mercury methylation in the changing environment: Recent advances in assessing microbial methylators and mercury bioavailability.Sci Total Environ. 2020 Apr 20;714:136827. doi: 10.1016/j.scitotenv.2020.136827. Epub 2020 Jan 22. Sci Total Environ. 2020. PMID: 32018974
-
Carbon Amendments Alter Microbial Community Structure and Net Mercury Methylation Potential in Sediments.Appl Environ Microbiol. 2018 Jan 17;84(3):e01049-17. doi: 10.1128/AEM.01049-17. Print 2018 Feb 1. Appl Environ Microbiol. 2018. PMID: 29150503 Free PMC article.
-
Mercury transformations in algae, plants, and animals: The occurrence, mechanisms, and gaps.Sci Total Environ. 2024 Feb 10;911:168690. doi: 10.1016/j.scitotenv.2023.168690. Epub 2023 Nov 23. Sci Total Environ. 2024. PMID: 38000748 Review.
-
Microbial Mercury Methylation in Aquatic Environments: A Critical Review of Published Field and Laboratory Studies.Environ Sci Technol. 2019 Jan 2;53(1):4-19. doi: 10.1021/acs.est.8b02709. Epub 2018 Dec 21. Environ Sci Technol. 2019. PMID: 30525497 Review.
Cited by
-
Overview of Methylation and Demethylation Mechanisms and Influencing Factors of Mercury in Water.Toxics. 2024 Sep 30;12(10):715. doi: 10.3390/toxics12100715. Toxics. 2024. PMID: 39453135 Free PMC article. Review.
References
-
- Adediran GA, Liem-Nguyen V, Song Y, Schaefer JK, Skyllberg U, Bjorn E (2019) Microbial biosynthesis of thiol compounds: implications for speciation, cellular uptake, and methylation of Hg(II). Environ Sci Technol 53(14):8187–8196. 10.1021/acs.est.9b01502 - PubMed
-
- An Y, Zhang R, Yang S, Wang Y, Lei Y, Peng S, Song L (2022) Microbial mercury methylation potential in a large-scale municipal solid waste landfill, China. Waste Manage 145:102–111. 10.1016/j.wasman.2022.04.038 - PubMed
-
- Avramescu ML, Yumvihoze E, Hintelmann H, Ridal J, Fortin D, Lean DR (2011) Biogeochemical factors influencing net mercury methylation in contaminated freshwater sediments from the St. Lawrence River in Cornwall, Ontario, Canada. Sci Total Environ 409(5):968–978. 10.1016/j.scitotenv.2010.11.016 - PubMed
-
- Azaroff A, Urriza MG, Gassie C, Monperrus M, Guyoneaud R (2020) Marine mercury-methylating microbial communities from coastal to Capbreton Canyon sediments (North Atlantic Ocean). Environ Pollut 262:114333. 10.1016/j.envpol.2020.114333 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

