Targeting autophagy overcomes cancer-intrinsic resistance to CAR-T immunotherapy in B-cell malignancies
- PMID: 38407943
- PMCID: PMC10958674
- DOI: 10.1002/cac2.12525
Targeting autophagy overcomes cancer-intrinsic resistance to CAR-T immunotherapy in B-cell malignancies
Abstract
Background: Chimeric antigen receptor T (CAR-T) therapy has substantially revolutionized the clinical outcomes of patients with hematologic malignancies, but the cancer-intrinsic mechanisms underlying resistance to CAR-T cells remain yet to be fully understood. This study aims to explore the molecular determinants of cancer cell sensitivity to CAR-T cell-mediated killing and to provide a better understanding of the underlying mechanisms and potential modulation to improve clinical efficacy.
Methods: The human whole-genome CRISPR/Cas9-based knockout screening was conducted to identify key genes that enable cancer cells to evade CD19 CAR-T-cell-mediated killing. The in vitro cytotoxicity assays and evaluation of tumor tissue and bone marrow specimens were further conducted to confirm the role of the key genes in cancer cell susceptibility to CAR-T cells. In addition, the specific mechanisms influencing CAR-T cell-mediated cancer clearance were elucidated in mouse and cellular models.
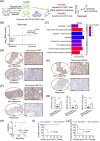
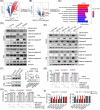
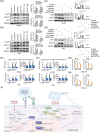
Results: The CRISPR/Cas9-based knockout screening showed that the enrichment of autophagy-related genes (ATG3, BECN1, and RB1CC1) provided protection of cancer cells from CD19 CAR-T cell-mediated cytotoxicity. These findings were further validated by in vitro cytotoxicity assays in cells with genetic and pharmacological inhibition of autophagy. Notably, higher expression of the three autophagy-related proteins in tumor samples was correlated with poorer responsiveness and worse survival in patients with relapsed/refractory B-cell lymphoma after CD19 CAR-T therapy. Bulk RNA sequencing analysis of bone marrow samples from B-cell leukemia patients also suggested the clinical relevance of autophagy to the therapeutic response and relapse after CD19 CAR-T cell therapy. Pharmacological inhibition of autophagy and knockout of RB1CC1 could dramatically sensitize tumor cells to CD19 CAR-T cell-mediated killing in mouse models of both B-cell leukemia and lymphoma. Moreover, our study revealed that cancer-intrinsic autophagy mediates evasion of CAR-T cells via the TNF-α-TNFR1 axis-mediated apoptosis and STAT1/IRF1-induced chemokine signaling activation.
Conclusions: These findings confirm that autophagy signaling in B-cell malignancies is essential for the effective cytotoxic function of CAR-T cells and thereby pave the way for the development of autophagy-targeting strategies to improve the clinical efficacy of CAR-T cell immunotherapy.
Keywords: Apoptosis; Autophagy; CAR‐T resistance; Chemotaxis; Immune evasion.
© 2024 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat‐sen University Cancer Center.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

