Detection of driver mutations and genomic signatures in endometrial cancers using artificial intelligence algorithms
- PMID: 38408048
- PMCID: PMC10896512
- DOI: 10.1371/journal.pone.0299114
Detection of driver mutations and genomic signatures in endometrial cancers using artificial intelligence algorithms
Abstract
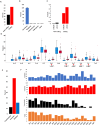
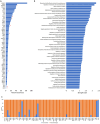
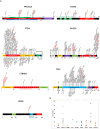
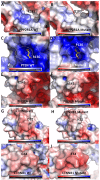
Analyzed endometrial cancer (EC) genomes have allowed for the identification of molecular signatures, which enable the classification, and sometimes prognostication, of these cancers. Artificial intelligence algorithms have facilitated the partitioning of mutations into driver and passenger based on a variety of parameters, including gene function and frequency of mutation. Here, we undertook an evaluation of EC cancer genomes deposited on the Catalogue of Somatic Mutations in Cancers (COSMIC), with the goal to classify all mutations as either driver or passenger. Our analysis showed that approximately 2.5% of all mutations are driver and cause cellular transformation and immortalization. We also characterized nucleotide level mutation signatures, gross chromosomal re-arrangements, and gene expression profiles. We observed that endometrial cancers show distinct nucleotide substitution and chromosomal re-arrangement signatures compared to other cancers. We also identified high expression levels of the CLDN18 claudin gene, which is involved in growth, survival, metastasis and proliferation. We then used in silico protein structure analysis to examine the effect of certain previously uncharacterized driver mutations on protein structure. We found that certain mutations in CTNNB1 and TP53 increase protein stability, which may contribute to cellular transformation. While our analysis retrieved previously classified mutations and genomic alterations, which is to be expected, this study also identified new signatures. Additionally, we show that artificial intelligence algorithms can be effectively leveraged to accurately predict key drivers of cancer. This analysis will expand our understanding of ECs and improve the molecular toolbox for classification, diagnosis, or potential treatment of these cancers.
Copyright: © 2024 Stan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Genomic Analysis of Uterine Lavage Fluid Detects Early Endometrial Cancers and Reveals a Prevalent Landscape of Driver Mutations in Women without Histopathologic Evidence of Cancer: A Prospective Cross-Sectional Study.PLoS Med. 2016 Dec 27;13(12):e1002206. doi: 10.1371/journal.pmed.1002206. eCollection 2016 Dec. PLoS Med. 2016. PMID: 28027320 Free PMC article.
-
Utility of a custom designed next generation DNA sequencing gene panel to molecularly classify endometrial cancers according to The Cancer Genome Atlas subgroups.BMC Med Genomics. 2020 Nov 30;13(1):179. doi: 10.1186/s12920-020-00824-8. BMC Med Genomics. 2020. PMID: 33256706 Free PMC article.
-
Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis.BMC Bioinformatics. 2014 Sep 19;15(1):308. doi: 10.1186/1471-2105-15-308. BMC Bioinformatics. 2014. PMID: 25236784 Free PMC article.
-
Identifying driver mutations from sequencing data of heterogeneous tumors in the era of personalized genome sequencing.Brief Bioinform. 2014 Mar;15(2):244-55. doi: 10.1093/bib/bbt042. Epub 2013 Jul 1. Brief Bioinform. 2014. PMID: 23818492 Review.
-
The Histomorphology to Molecular Transition: Exploring the Genomic Landscape of Poorly Differentiated Epithelial Endometrial Cancers.Cells. 2025 Mar 5;14(5):382. doi: 10.3390/cells14050382. Cells. 2025. PMID: 40072110 Free PMC article. Review.
Cited by
-
In silico analysis of several frequent SLX4 mutations appearing in human cancers.MicroPubl Biol. 2024 May 17;2024:10.17912/micropub.biology.001216. doi: 10.17912/micropub.biology.001216. eCollection 2024. MicroPubl Biol. 2024. PMID: 38828439 Free PMC article.
-
From Genes to Clinical Practice: Exploring the Genomic Underpinnings of Endometrial Cancer.Cancers (Basel). 2025 Jan 20;17(2):320. doi: 10.3390/cancers17020320. Cancers (Basel). 2025. PMID: 39858102 Free PMC article. Review.
-
CD73 restrains mutant β-catenin oncogenic activity in endometrial carcinomas.bioRxiv [Preprint]. 2024 Nov 18:2024.11.18.624183. doi: 10.1101/2024.11.18.624183. bioRxiv. 2024. PMID: 39605508 Free PMC article. Preprint.
-
An Overview of Artificial Intelligence in Gynaecological Pathology Diagnostics.Cancers (Basel). 2025 Apr 16;17(8):1343. doi: 10.3390/cancers17081343. Cancers (Basel). 2025. PMID: 40282519 Free PMC article. Review.
-
Epigenetics of Endometrial Cancer: The Role of Chromatin Modifications and Medicolegal Implications.Int J Mol Sci. 2025 Jul 29;26(15):7306. doi: 10.3390/ijms26157306. Int J Mol Sci. 2025. PMID: 40806438 Free PMC article. Review.
References
-
- Centers for Disease Control and Prevention. (2023). Gynecological Cancers. https://www.cdc.gov/cancer/gynecologic/index.htm.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

