Histone deficiency and hypoacetylation in the aging retinal pigment epithelium
- PMID: 38408164
- PMCID: PMC11113634
- DOI: 10.1111/acel.14108
Histone deficiency and hypoacetylation in the aging retinal pigment epithelium
Abstract
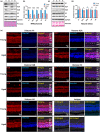
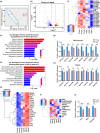
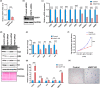
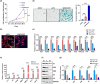
Histones serve as a major carrier of epigenetic information in the form of post-translational modifications which are vital for controlling gene expression, maintaining cell identity, and ensuring proper cellular function. Loss of histones in the aging genome can drastically impact the epigenetic landscape of the cell leading to altered chromatin structure and changes in gene expression profiles. In this study, we investigated the impact of age-related changes on histone levels and histone acetylation in the retinal pigment epithelium (RPE) and retina of mice. We observed a global reduction of histones H1, H2A, H2B, H3, and H4 in aged RPE/choroid but not in the neural retina. Transcriptomic analyses revealed significant downregulation of histones in aged RPE/choroid including crucial elements of the histone locus body (HLB) complex involved in histone pre-mRNA processing. Knockdown of HINFP, a key HLB component, in human RPE cells induced histone loss, senescence, and the upregulation of senescence-associated secretory phenotype (SASP) markers. Replicative senescence and chronological aging in human RPE cells similarly resulted in progressive histone loss and acquisition of the SASP. Immunostaining of human retina sections revealed histone loss in RPE with age. Acetyl-histone profiling in aged mouse RPE/choroid revealed a specific molecular signature with loss of global acetyl-histone levels, including H3K14ac, H3K56ac, and H4K16ac marks. These findings strongly demonstrate histone loss as a unique feature of RPE aging and provide critical insights into the potential mechanisms linking histone dynamics, cellular senescence, and aging.
Keywords: HINFP; aging; epigenetics; histone acetylation; histones; replicative senescence; retinal pigment epithelium.
© 2024 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Deciphering age-related transcriptomic changes in the mouse retinal pigment epithelium.Aging (Albany NY). 2025 Mar 4;17(3):657-684. doi: 10.18632/aging.206219. Epub 2025 Mar 4. Aging (Albany NY). 2025. PMID: 40042930 Free PMC article.
-
Histone modifications contribute to cellular replicative and hydrogen peroxide-induced premature senescence in human embryonic lung fibroblasts.Free Radic Res. 2014 May;48(5):550-9. doi: 10.3109/10715762.2014.893580. Epub 2014 Mar 10. Free Radic Res. 2014. PMID: 24528089
-
Aging induces cell loss and a decline in phagosome processing in the mouse retinal pigment epithelium.Neurobiol Aging. 2023 Aug;128:1-16. doi: 10.1016/j.neurobiolaging.2023.03.003. Epub 2023 Mar 13. Neurobiol Aging. 2023. PMID: 37130462
-
Unraveling Histone Loss in Aging and Senescence.Cells. 2024 Feb 9;13(4):320. doi: 10.3390/cells13040320. Cells. 2024. PMID: 38391933 Free PMC article. Review.
-
Epigenetic Regulation of Cellular Senescence.Cells. 2022 Feb 15;11(4):672. doi: 10.3390/cells11040672. Cells. 2022. PMID: 35203320 Free PMC article. Review.
Cited by
-
Epigenetic Modifications in the Retinal Pigment Epithelium of the Eye During RPE-Related Regeneration or Retinal Diseases in Vertebrates.Biomedicines. 2025 Jun 25;13(7):1552. doi: 10.3390/biomedicines13071552. Biomedicines. 2025. PMID: 40722628 Free PMC article. Review.
-
Deciphering age-related transcriptomic changes in the mouse retinal pigment epithelium.Aging (Albany NY). 2025 Mar 4;17(3):657-684. doi: 10.18632/aging.206219. Epub 2025 Mar 4. Aging (Albany NY). 2025. PMID: 40042930 Free PMC article.
-
Epigenetic Alterations in Age-Related Macular Degeneration: Mechanisms and Implications.Int J Mol Sci. 2025 Aug 6;26(15):7601. doi: 10.3390/ijms26157601. Int J Mol Sci. 2025. PMID: 40806731 Free PMC article. Review.
-
Protein Acetylation in Age-Related Macular Degeneration: Mechanisms, Roles, and Therapeutic Perspectives.Invest Ophthalmol Vis Sci. 2025 May 1;66(5):30. doi: 10.1167/iovs.66.5.30. Invest Ophthalmol Vis Sci. 2025. PMID: 40402519 Free PMC article. Review.
-
Aere perennius: how chromatin fidelity is maintained and lost in disease.NAR Mol Med. 2025 Jul 22;2(3):ugaf026. doi: 10.1093/narmme/ugaf026. eCollection 2025 Jul. NAR Mol Med. 2025. PMID: 40809411 Free PMC article. Review.
References
-
- Barcaroli, D. , Bongiorno‐Borbone, L. , Terrinoni, A. , Hofmann, T. G. , Rossi, M. , Knight, R. A. , Matera, A. G. , Melino, G. , & De Laurenzi, V. (2006). FLASH is required for histone transcription and S‐phase progression. Proceedings of the National Academy of Sciences of the United States of America, 103(40), 14808–14812. 10.1073/pnas.0604227103 - DOI - PMC - PubMed
-
- Berdasco, M. , Gomez, A. , Rubio, M. J. , Catala‐Mora, J. , Zanon‐Moreno, V. , Lopez, M. , Hernández, C. , Yoshida, S. , Nakama, T. , Ishikawa, K. , Ishibashi, T. , Boubekeur, A. M. , Louhibi, L. , Pujana, M. A. , Sayols, S. , Setien, F. , Corella, D. , de Torres, C. , Parareda, A. , … Esteller, M. (2017). DNA methylomes reveal biological networks involved in human eye development, functions and associated disorders. Scientific Reports, 7(1), 11762. 10.1038/s41598-017-12084-1 - DOI - PMC - PubMed
-
- Blake, J. A. , Baldarelli, R. , Kadin, J. A. , Richardson, J. E. , Smith, C. L. , Bult, C. J. , & Mouse Genome Database Group . (2021). Mouse genome database (MGD): Knowledgebase for mouse‐human comparative biology. Nucleic Acids Research, 49(D1), D981–D987. 10.1093/nar/gkaa1083 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

