A patterned human neural tube model using microfluidic gradients
- PMID: 38408487
- PMCID: PMC11006583
- DOI: 10.1038/s41586-024-07204-7
A patterned human neural tube model using microfluidic gradients
Abstract
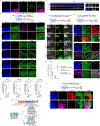
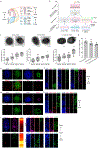
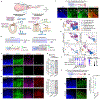
The human nervous system is a highly complex but organized organ. The foundation of its complexity and organization is laid down during regional patterning of the neural tube, the embryonic precursor to the human nervous system. Historically, studies of neural tube patterning have relied on animal models to uncover underlying principles. Recently, models of neurodevelopment based on human pluripotent stem cells, including neural organoids1-5 and bioengineered neural tube development models6-10, have emerged. However, such models fail to recapitulate neural patterning along both rostral-caudal and dorsal-ventral axes in a three-dimensional tubular geometry, a hallmark of neural tube development. Here we report a human pluripotent stem cell-based, microfluidic neural tube-like structure, the development of which recapitulates several crucial aspects of neural patterning in brain and spinal cord regions and along rostral-caudal and dorsal-ventral axes. This structure was utilized for studying neuronal lineage development, which revealed pre-patterning of axial identities of neural crest progenitors and functional roles of neuromesodermal progenitors and the caudal gene CDX2 in spinal cord and trunk neural crest development. We further developed dorsal-ventral patterned microfluidic forebrain-like structures with spatially segregated dorsal and ventral regions and layered apicobasal cellular organizations that mimic development of the human forebrain pallium and subpallium, respectively. Together, these microfluidics-based neurodevelopment models provide three-dimensional lumenal tissue architectures with in vivo-like spatiotemporal cell differentiation and organization, which will facilitate the study of human neurodevelopment and disease.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures













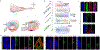


References
References for Main Text
-
- Faustino Martins J-M et al. Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell 26, 172–186.e176 (2020). - PubMed
References for Methods section
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

