Altered hippocampal neurogenesis in a mouse model of autism revealed by genetic polymorphisms and by atypical development of newborn neurons
- PMID: 38409172
- PMCID: PMC10897317
- DOI: 10.1038/s41598-024-53614-y
Altered hippocampal neurogenesis in a mouse model of autism revealed by genetic polymorphisms and by atypical development of newborn neurons
Abstract
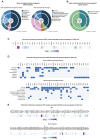
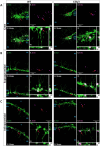
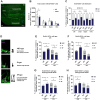
Individuals with autism spectrum disorder (ASD) often exhibit atypical hippocampal anatomy and connectivity throughout their lifespan, potentially linked to alterations in the neurogenic process within the hippocampus. In this study, we performed an in-silico analysis to identify single-nucleotide polymorphisms (SNPs) in genes relevant to adult neurogenesis in the C58/J model of idiopathic autism. We found coding non-synonymous (Cn) SNPs in 33 genes involved in the adult neurogenic process, as well as in 142 genes associated with the signature genetic profile of neural stem cells (NSC) and neural progenitors. Based on the potential alterations in adult neurogenesis predicted by the in-silico analysis, we evaluated the number and distribution of newborn neurons in the dentate gyrus (DG) of young adult C58/J mice. We found a reduced number of newborn cells in the whole DG, a higher proportion of early neuroblasts in the subgranular layer (SGZ), and a lower proportion of neuroblasts with morphological maturation signs in the granule cell layer (GCL) of the DG compared to C57BL/6J mice. The observed changes may be associated with a delay in the maturation trajectory of newborn neurons in the C58/J strain, linked to the Cn SNPs in genes involved in adult hippocampal neurogenesis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Single-nucleotide polymorphism analysis accurately predicts multiple impairments in hippocampal activity and memory performance in a murine model of idiopathic autism.Sci Rep. 2025 Jan 4;15(1):749. doi: 10.1038/s41598-024-84521-x. Sci Rep. 2025. PMID: 39755808 Free PMC article.
-
Cux2 activity defines a subpopulation of perinatal neurogenic progenitors in the hippocampus.Hippocampus. 2015 Feb;25(2):253-67. doi: 10.1002/hipo.22370. Epub 2014 Oct 3. Hippocampus. 2015. PMID: 25252086 Free PMC article.
-
CAPS2 deficiency affects environmental enrichment-induced adult neurogenesis and differentiation/survival of newborn neurons in the hippocampal dentate gyrus.Neurosci Lett. 2017 Nov 20;661:121-125. doi: 10.1016/j.neulet.2017.09.047. Epub 2017 Sep 27. Neurosci Lett. 2017. PMID: 28963059
-
How to make a hippocampal dentate gyrus granule neuron.Development. 2014 Jun;141(12):2366-75. doi: 10.1242/dev.096776. Development. 2014. PMID: 24917496 Review.
-
Implication of Hippocampal Neurogenesis in Autism Spectrum Disorder: Pathogenesis and Therapeutic Implications.Curr Neuropharmacol. 2023;21(11):2266-2282. doi: 10.2174/1570159X21666221220155455. Curr Neuropharmacol. 2023. PMID: 36545727 Free PMC article. Review.
Cited by
-
Single-nucleotide polymorphism analysis accurately predicts multiple impairments in hippocampal activity and memory performance in a murine model of idiopathic autism.Sci Rep. 2025 Jan 4;15(1):749. doi: 10.1038/s41598-024-84521-x. Sci Rep. 2025. PMID: 39755808 Free PMC article.
-
Adult neurogenesis in the ventral hippocampus decreased among animal models of neurodevelopmental disorders.Front Neural Circuits. 2024 Dec 23;18:1504191. doi: 10.3389/fncir.2024.1504191. eCollection 2024. Front Neural Circuits. 2024. PMID: 39764171 Free PMC article.
References
-
- American Psychiatric Association . Diagnostic And Statistical Manual Of Mental Disorders, Fifth Edition, Text Revision (DSM-5-TR) American Psychiatric Association Publishing; 2022.
-
- World Health Organization. Autism. Fact sheets: Autismhttps://www.who.int/news-room/fact-sheets/detail/autism-spectrum-disorders (2023).
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

