Bright and stable monomeric green fluorescent protein derived from StayGold
- PMID: 38409224
- PMCID: PMC11852770
- DOI: 10.1038/s41592-024-02203-y
Bright and stable monomeric green fluorescent protein derived from StayGold
Abstract
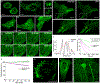
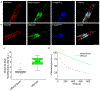
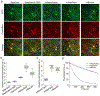
The high brightness and photostability of the green fluorescent protein StayGold make it a particularly attractive probe for long-term live-cell imaging; however, its dimeric nature precludes its application as a fluorescent tag for some proteins. Here, we report the development and crystal structures of a monomeric variant of StayGold, named mBaoJin, which preserves the beneficial properties of its precursor, while serving as a tag for structural proteins and membranes. Systematic benchmarking of mBaoJin against popular green fluorescent proteins and other recently introduced monomeric and pseudomonomeric derivatives of StayGold established mBaoJin as a bright and photostable fluorescent protein, exhibiting rapid maturation and high pH/chemical stability. mBaoJin was also demonstrated for super-resolution, long-term live-cell imaging and expansion microscopy. We further showed the applicability of mBaoJin for neuronal labeling in model organisms, including Caenorhabditis elegans and mice.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
K.D.P. is the co-founder of a company that pursues commercial applications of expansion microscopy and is listed as an inventor on several patent applications concerning the development of new expansion microscopy methods. All other authors have no competing interests.
Figures


References
-
- Jacquemet G, Carisey AF, Hamidi H, Henriques R & Leterrier C The cell biologist’s guide to super-resolution microscopy. J. Cell Sci 133, (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

