Targeting PHGDH reverses the immunosuppressive phenotype of tumor-associated macrophages through α-ketoglutarate and mTORC1 signaling
- PMID: 38409249
- PMCID: PMC11061172
- DOI: 10.1038/s41423-024-01134-0
Targeting PHGDH reverses the immunosuppressive phenotype of tumor-associated macrophages through α-ketoglutarate and mTORC1 signaling
Abstract
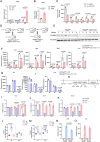
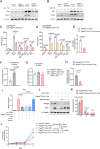
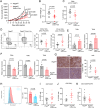
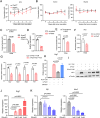
Phosphoglycerate dehydrogenase (PHGDH) has emerged as a crucial factor in macromolecule synthesis, neutralizing oxidative stress, and regulating methylation reactions in cancer cells, lymphocytes, and endothelial cells. However, the role of PHGDH in tumor-associated macrophages (TAMs) is poorly understood. Here, we found that the T helper 2 (Th2) cytokine interleukin-4 and tumor-conditioned media upregulate the expression of PHGDH in macrophages and promote immunosuppressive M2 macrophage activation and proliferation. Loss of PHGDH disrupts cellular metabolism and mitochondrial respiration, which are essential for immunosuppressive macrophages. Mechanistically, PHGDH-mediated serine biosynthesis promotes α-ketoglutarate production, which activates mTORC1 signaling and contributes to the maintenance of an M2-like macrophage phenotype in the tumor microenvironment. Genetic ablation of PHGDH in macrophages from tumor-bearing mice results in attenuated tumor growth, reduced TAM infiltration, a phenotypic shift of M2-like TAMs toward an M1-like phenotype, downregulated PD-L1 expression and enhanced antitumor T-cell immunity. Our study provides a strong basis for further exploration of PHGDH as a potential target to counteract TAM-mediated immunosuppression and hinder tumor progression.
Keywords: PHGDH; de novo serine synthesis; mTORC1; protumorigenic; tumor-associated macrophages, metabolomics; α-ketoglutarate.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Kloosterman DJ, Akkari L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell. 2023;186:1627–51. - PubMed
-
- Dehne N, Mora J, Namgaladze D, Weigert A, Brüne B. Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol. 2017;35:12–19. - PubMed
-
- Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17:887–904. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P32600/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- P 34023/FWF_/Austrian Science Fund FWF/Austria
- P 30857/FWF_/Austrian Science Fund FWF/Austria
- ESP87/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- P 32600/FWF_/Austrian Science Fund FWF/Austria
- 201908320480/CSC | Chinese Government Scholarship
- 201806500012/CSC | Chinese Government Scholarship
- LS18-058/Vienna Science and Technology Fund (Wiener Wissenschafts-, Forschungs- und Technologiefonds)
- ESP 87/FWF_/Austrian Science Fund FWF/Austria
- P34023-B/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

