Evidence review and considerations for use of first line genome sequencing to diagnose rare genetic disorders
- PMID: 38409289
- PMCID: PMC10897481
- DOI: 10.1038/s41525-024-00396-x
Evidence review and considerations for use of first line genome sequencing to diagnose rare genetic disorders
Abstract
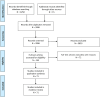
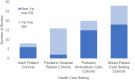
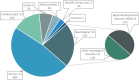
Early use of genome sequencing (GS) in the diagnostic odyssey can reduce suffering and improve care, but questions remain about which patient populations are most amenable to GS as a first-line diagnostic test. To address this, the Medical Genome Initiative conducted a literature review to identify appropriate clinical indications for GS. Studies published from January 2011 to August 2022 that reported on the diagnostic yield (DY) or clinical utility of GS were included. An exploratory meta-analysis using a random effects model evaluated DY based on cohort size and diagnosed cases per cohort. Seventy-one studies met inclusion criteria, comprising over 13,000 patients who received GS in one of the following settings: hospitalized pediatric patients, pediatric outpatients, adult outpatients, or mixed. GS was the first-line test in 38% (27/71). The unweighted mean DY of first-line GS was 45% (12-73%), 33% (6-86%) in cohorts with prior genetic testing, and 33% (9-60%) in exome-negative cohorts. Clinical utility was reported in 81% of first-line GS studies in hospitalized pediatric patients. Changes in management varied by cohort and underlying molecular diagnosis (24-100%). To develop evidence-informed points to consider, the quality of all 71 studies was assessed using modified American College of Radiology (ACR) criteria, with five core points to consider developed, including recommendations for use of GS in the N/PICU, in lieu of sequential testing and when disorders with substantial allelic heterogeneity are suspected. Future large and controlled studies in the pediatric and adult populations may support further refinement of these recommendations.
© 2024. The Author(s).
Conflict of interest statement
Stacie L. Taylor and Ryan J. Taft are current shareholders and employees of Illumina Inc. The remaining authors declare no competing interests.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

