Pooled effector library screening in protoplasts rapidly identifies novel Avr genes
- PMID: 38409291
- PMCID: PMC11035141
- DOI: 10.1038/s41477-024-01641-y
Pooled effector library screening in protoplasts rapidly identifies novel Avr genes
Abstract
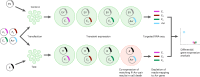
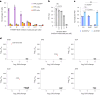
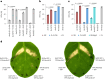
Crop breeding for durable disease resistance is challenging due to the rapid evolution of pathogen virulence. While progress in resistance (R) gene cloning and stacking has accelerated in recent years1-3, the identification of corresponding avirulence (Avr) genes in many pathogens is hampered by the lack of high-throughput screening options. To address this technology gap, we developed a platform for pooled library screening in plant protoplasts to allow rapid identification of interacting R-Avr pairs. We validated this platform by isolating known and novel Avr genes from wheat stem rust (Puccinia graminis f. sp. tritici) after screening a designed library of putative effectors against individual R genes. Rapid Avr gene identification provides molecular tools to understand and track pathogen virulence evolution via genotype surveillance, which in turn will lead to optimized R gene stacking and deployment strategies. This platform should be broadly applicable to many crop pathogens and could potentially be adapted for screening genes involved in other protoplast-selectable traits.
© 2024. The Author(s).
Conflict of interest statement
The protoplast screening platform described here is the subject of a patent application filed by CSIRO, Australian Provisional Patent Application No. 2022903442.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

