A homoeostatic switch causing glycerol-3-phosphate and phosphoethanolamine accumulation triggers senescence by rewiring lipid metabolism
- PMID: 38409325
- PMCID: PMC10896726
- DOI: 10.1038/s42255-023-00972-y
A homoeostatic switch causing glycerol-3-phosphate and phosphoethanolamine accumulation triggers senescence by rewiring lipid metabolism
Abstract
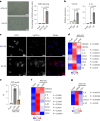
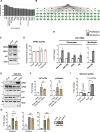
Cellular senescence affects many physiological and pathological processes and is characterized by durable cell cycle arrest, an inflammatory secretory phenotype and metabolic reprogramming. Here, by using dynamic transcriptome and metabolome profiling in human fibroblasts with different subtypes of senescence, we show that a homoeostatic switch that results in glycerol-3-phosphate (G3P) and phosphoethanolamine (pEtN) accumulation links lipid metabolism to the senescence gene expression programme. Mechanistically, p53-dependent glycerol kinase activation and post-translational inactivation of phosphate cytidylyltransferase 2, ethanolamine regulate this metabolic switch, which promotes triglyceride accumulation in lipid droplets and induces the senescence gene expression programme. Conversely, G3P phosphatase and ethanolamine-phosphate phospho-lyase-based scavenging of G3P and pEtN acts in a senomorphic way by reducing G3P and pEtN accumulation. Collectively, our study ties G3P and pEtN accumulation to controlling lipid droplet biogenesis and phospholipid flux in senescent cells, providing a potential therapeutic avenue for targeting senescence and related pathophysiology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
















References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

