Decanoylcarnitine Inhibits Triple-Negative Breast Cancer Progression via Mmp9 in an Intermittent Fasting Obesity Mouse
- PMID: 38409962
- PMCID: PMC10898300
- DOI: 10.1177/15330338241233443
Decanoylcarnitine Inhibits Triple-Negative Breast Cancer Progression via Mmp9 in an Intermittent Fasting Obesity Mouse
Abstract
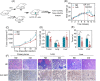
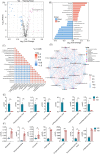
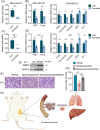
Purpose: Treatment of triple-negative breast cancer (TNBC) remains challenging. Intermittent fasting (IF) has emerged as a promising approach to improve metabolic health of various metabolic disorders. Clinical studies indicate IF is essential for TNBC progression. However, the molecular mechanisms underlying metabolic remodeling in regulating IF and TNBC progression are still unclear. Methods: In this study, we utilized a robust mouse model of TNBC and exposed subjects to a high-fat diet (HFD) with IF to explore its impact on the metabolic reprogramming linked to cancer progression. To identify crucial serum metabolites and signaling events, we utilized targeted metabolomics and RNA sequencing (RNA-seq). Furthermore, we conducted immunoblotting, real-time quantitative polymerase chain reaction (RT-qPCR), cell migration assays, lentivirus-mediated Mmp9 overexpression, and Mmp9 inhibitor experiments to elucidate the role of decanoylcarnitine/Mmp9 in TNBC cell migration. Results: Our observations indicate that IF exerts notable inhibitory effects on both the proliferation and cancer metastasis. Utilizing targeted metabolomics and RNA-seq, we initially identified pivotal serum metabolites and signaling events in the progression of TNBC. Among the 349 serum metabolites identified, decanoylcarnitine was picked out to inhibit TNBC cell proliferation and migration. RNA-seq analysis of TNBC cells treated with decanoylcarnitine revealed its suppressive effects on extracellular matrix-related protein components, with a notable reduction observed in Mmp9. Further investigations confirmed that decanoylcarnitine could inhibit Mmp9 expression in TNBC cells, primary tumors, lung, and liver metastasis tissues. Mmp9 overexpression abolished the inhibitory effect of decanoylcarnitine on cell migration. Conclusion: This study pioneers the exploration of IF intervention and the role of decanoylcarnitine/Mmp9 in the progression of TNBC in obese mice, enhancing our comprehension of the potential roles of various dietary patterns in the process of cancer treatment.
Keywords: cell migration; decanoylcarnitine; integrated multiomic analysis; intermittent fasting; triple-negative breast cancer.
Conflict of interest statement
Declaration of Conflicting InterestsThe authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures




References
-
- Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer-expanded options, evolving needs. Nat Rev Clin Oncol. 2022;19(2):91-113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

