Characterization of pathogenic microbiome on removable prostheses with different levels of cleanliness using 2bRAD-M metagenomic sequencing
- PMID: 38410192
- PMCID: PMC10896157
- DOI: 10.1080/20002297.2024.2317059
Characterization of pathogenic microbiome on removable prostheses with different levels of cleanliness using 2bRAD-M metagenomic sequencing
Abstract
Background: The microbiomes on the surface of unclean removable prostheses are complex and yet largely underexplored using metagenomic sequencing technology.
Objectives: To characterize the microbiome of removable prostheses with different levels of cleanliness using Type IIB Restriction-site Associated DNA for Microbiome (2bRAD-M) sequencing and compare the Microbial Index of Pathogenic Bacteria (MIP) between clean and unclean prostheses.
Materials and methods: Ninety-seven removable prostheses were classified into 'clean' and 'unclean' groups. All prosthesis plaque samples underwent 2bRAD metagenomic sequencing to characterize the species-resolved microbial composition. MIPs for clean and unclean prostheses were calculated based on the sum of the relative abundance of pathogenic bacteria in a microbiome using a reference database that contains opportunistic pathogenic bacteria and disease-associated information.
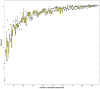
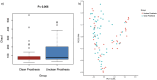
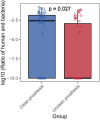
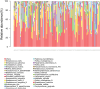
Results: Beta diversity analyses based on Jaccard qualitative and Bray-Curtis quantitative distance matrices identified significant differences between the two groups (p < 0.05). There was a significant enrichment of many pathogenic bacteria in the unclean prosthesis group. The MIP for unclean prostheses (0.47 ± 0.25) was significantly higher than for clean prostheses (0.37 ± 0.29), p = 0.029.
Conclusions: The microbial community of plaque samples from 'unclean' prostheses demonstrated compositional differences compared with 'clean' prostheses. In addition, the pathogenic microbiome in the 'unclean' versus 'clean' group differed.
Keywords: Removable prosthesis; metagenomics; microbial index; pathogenic bacteria; prosthesis cleanliness.
Plain language summary
The pathogenic microbiome in the unclean removable prosthesis group tends to be more abundant than that of the clean counterpart among participants with the majority being elders attending a teaching hospital. This finding is worrying because a general decline in systemic health among community-dwelling elders may predispose them to life-threatening diseases.By understanding the characteristics of the microbiome of removable prostheses with different levels of cleanliness and the related microbial-infection risks after a comprehensive whole metagenomic sequencing, appropriate prosthesis hygiene care should be emphasized.This study introduced a comprehensive and novel method of microbiological investigation of plaque using 2bRAD-M. The taxonomic profile of the microbiome of whole genomes was demonstrated and resolved at the species-level taxonomy for prosthesis biofilms. The biggest advantage of this method included overcoming the DNA sample problems particularly the low-biomass nature of ‘clean’ prosthesis plaque samples. The findings will add to our understanding of the microbiological aspect of removable prosthesis plaque.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
LinkOut - more resources
Full Text Sources
