Major primary bile salts repress Salmonella enterica serovar Typhimurium invasiveness partly via the efflux regulatory locus ramRA
- PMID: 38410385
- PMCID: PMC10895713
- DOI: 10.3389/fmicb.2024.1338261
Major primary bile salts repress Salmonella enterica serovar Typhimurium invasiveness partly via the efflux regulatory locus ramRA
Abstract
Bile represses Salmonella enterica serovar Typhimurium (S. Typhimurium) intestinal cell invasion, but it remains unclear which bile components and mechanisms are implicated. Previous studies reported that bile inhibits the RamR binding to the ramA promoter, resulting in ramA increased transcription, and that ramA overexpression is associated to decreased expression of type III secretion system 1 (TTSS-1) invasion genes and to impaired intestinal cell invasiveness in S. Typhimurium. In this study, we assessed the possible involvement of the ramRA multidrug efflux regulatory locus and individual bile salts in the bile-mediated repression of S. Typhimurium invasion, using Caco-2 intestinal epithelial cells and S. Typhimurium strain ATCC 14028s. Our results indicate that (i) major primary bile salts, chenodeoxycholate and its conjugated-derivative salts, cholate, and deoxycholate, activate ramA transcription in a RamR-dependent manner, and (ii) it results in repression of hilA, encoding the master activator of TTSS-1 genes, and as a consequence in the repression of cellular invasiveness. On the other hand, crude ox bile extract and cholate were also shown to repress the transcription of hilA independently of RamR, and to inhibit cell invasion independently of ramRA. Altogether, these data suggest that bile-mediated repression of S. Typhimurium invasion occurs through pleiotropic effects involving partly ramRA, as well as other unknown regulatory pathways. Bile components other than the bile salts used in this study might also participate in this phenomenon.
Keywords: RamR; Salmonella; Typhimurium; bile; intestinal; invasion; ramA; regulation.
Copyright © 2024 Giraud, Baucheron, Foubert, Doublet, Nishino and Cloeckaert.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
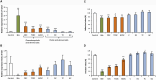
 crude bile extract,
crude bile extract,  chenodeoxycholic acid-derived salts (CDC, chenodeoxycholate; TCDC, taurochenodeoxycholate; GCDC, glycochenodeoxycholate) and
chenodeoxycholic acid-derived salts (CDC, chenodeoxycholate; TCDC, taurochenodeoxycholate; GCDC, glycochenodeoxycholate) and  cholic acid-derived salts (C, cholate; DC, deoxycholate; TC, taurocholate; GC, glycocholate).
cholic acid-derived salts (C, cholate; DC, deoxycholate; TC, taurocholate; GC, glycocholate).
 crude bile extract,
crude bile extract,  chenodeoxycholate (CDC),
chenodeoxycholate (CDC),  cholate (C), and
cholate (C), and  deoxycholate (DC). Asterisks indicate significant differences (NS, non-significant; *P < 0.05, **P < 0.01, ***P < 0.001). ND, not determined.
deoxycholate (DC). Asterisks indicate significant differences (NS, non-significant; *P < 0.05, **P < 0.01, ***P < 0.001). ND, not determined.
 , +) of crude bile extract at 25.6 g/L, of the WT S. Typhimurium strain 14028s and its ramRA::kan deletion mutant complemented with a pramA plasmid. Bars indicate the percentage of attached/internalized bacteria ± standard error of the mean. Asterisks indicate significant differences (***P < 0.001).
, +) of crude bile extract at 25.6 g/L, of the WT S. Typhimurium strain 14028s and its ramRA::kan deletion mutant complemented with a pramA plasmid. Bars indicate the percentage of attached/internalized bacteria ± standard error of the mean. Asterisks indicate significant differences (***P < 0.001).
Similar articles
-
Bile-mediated activation of the acrAB and tolC multidrug efflux genes occurs mainly through transcriptional derepression of ramA in Salmonella enterica serovar Typhimurium.J Antimicrob Chemother. 2014 Sep;69(9):2400-6. doi: 10.1093/jac/dku140. Epub 2014 May 9. J Antimicrob Chemother. 2014. PMID: 24816212
-
Effects of natural mutations in the ramRA locus on invasiveness of epidemic fluoroquinolone-resistant Salmonella enterica serovar Typhimurium isolates.J Infect Dis. 2013 Mar 1;207(5):794-802. doi: 10.1093/infdis/jis755. Epub 2012 Dec 10. J Infect Dis. 2013. PMID: 23230062
-
ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium.Antimicrob Agents Chemother. 2008 Jul;52(7):2428-34. doi: 10.1128/AAC.00084-08. Epub 2008 Apr 28. Antimicrob Agents Chemother. 2008. PMID: 18443112 Free PMC article.
-
Binding of the RamR repressor to wild-type and mutated promoters of the RamA gene involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium.Antimicrob Agents Chemother. 2012 Feb;56(2):942-8. doi: 10.1128/AAC.05444-11. Epub 2011 Nov 28. Antimicrob Agents Chemother. 2012. PMID: 22123696 Free PMC article.
-
ramR is not involved in the regulation of ramA associated antibiotic resistance in Salmonella enterica serovar Typhi.Microb Pathog. 2017 Oct;111:198-202. doi: 10.1016/j.micpath.2017.08.052. Epub 2017 Sep 5. Microb Pathog. 2017. PMID: 28882783
Cited by
-
Comparative genomic and antimicrobial resistance profiles of Salmonella strains isolated from pork and human sources in Sichuan, China.Front Microbiol. 2025 Mar 3;16:1515576. doi: 10.3389/fmicb.2025.1515576. eCollection 2025. Front Microbiol. 2025. PMID: 40099182 Free PMC article.
References
LinkOut - more resources
Full Text Sources

