Elevated levels of cell-free NKG2D-ligands modulate NKG2D surface expression and compromise NK cell function in severe COVID-19 disease
- PMID: 38410511
- PMCID: PMC10895954
- DOI: 10.3389/fimmu.2024.1273942
Elevated levels of cell-free NKG2D-ligands modulate NKG2D surface expression and compromise NK cell function in severe COVID-19 disease
Abstract
Introduction: It is now clear that coronavirus disease 19 (COVID-19) severity is associated with a dysregulated immune response, but the relative contributions of different immune cells is still not fully understood. SARS CoV-2 infection triggers marked changes in NK cell populations, but there are contradictory reports as to whether these effector lymphocytes play a protective or pathogenic role in immunity to SARS-CoV-2.
Methods: To address this question we have analysed differences in the phenotype and function of NK cells in SARS-CoV-2 infected individuals who developed either very mild, or life-threatening COVID-19 disease.
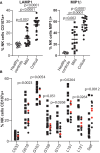
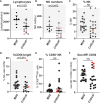
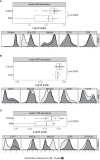
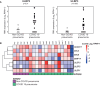
Results: Although NK cells from patients with severe disease appeared more activated and the frequency of adaptive NK cells was increased, they were less potent mediators of ADCC than NK cells from patients with mild disease. Further analysis of peripheral blood NK cells in these patients revealed that a population of NK cells that had lost expression of the activating receptor NKG2D were a feature of patients with severe disease and this correlated with elevated levels of cell free NKG2D ligands, especially ULBP2 and ULBP3 in the plasma of critically ill patients. In vitro, culture in NKG2DL containing patient sera reduced the ADCC function of healthy donor NK cells and this could be blocked by NKG2DL-specific antibodies.
Discussion: These observations of reduced NK function in severe disease are consistent with the hypothesis that defects in immune surveillance by NK cells permit higher levels of viral replication, rather than that aberrant NK cell function contributes to immune system dysregulation and immunopathogenicity.
Keywords: ADCC - antibody dependent cellular cytotoxicity; COVID-19; NKG2D (natural killer group 2 member D); natural killer cells; soluble NKG2D ligands.
Copyright © 2024 Fernández-Soto, García-Jiménez, Casasnovas, Valés-Gómez and Reyburn.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
Soluble ligands for the NKG2D receptor are released during HIV-1 infection and impair NKG2D expression and cytotoxicity of NK cells.FASEB J. 2013 Jun;27(6):2440-50. doi: 10.1096/fj.12-223057. Epub 2013 Feb 8. FASEB J. 2013. PMID: 23395909
-
An Fc-optimized NKG2D-immunoglobulin G fusion protein for induction of natural killer cell reactivity against leukemia.Int J Cancer. 2015 Mar 1;136(5):1073-84. doi: 10.1002/ijc.29083. Epub 2014 Jul 28. Int J Cancer. 2015. PMID: 25046567
-
SARS-CoV-2 escapes direct NK cell killing through Nsp1-mediated downregulation of ligands for NKG2D.Cell Rep. 2022 Dec 27;41(13):111892. doi: 10.1016/j.celrep.2022.111892. Epub 2022 Dec 12. Cell Rep. 2022. PMID: 36543165 Free PMC article.
-
The NKG2D/NKG2DL Axis in the Crosstalk Between Lymphoid and Myeloid Cells in Health and Disease.Front Immunol. 2018 Apr 23;9:827. doi: 10.3389/fimmu.2018.00827. eCollection 2018. Front Immunol. 2018. PMID: 29740438 Free PMC article. Review.
-
Targeting NKG2D/NKG2DL axis in multiple myeloma therapy.Cytokine Growth Factor Rev. 2024 Apr;76:1-11. doi: 10.1016/j.cytogfr.2024.02.001. Epub 2024 Feb 15. Cytokine Growth Factor Rev. 2024. PMID: 38378397 Review.
Cited by
-
NK cell based immunotherapy against oral squamous cell carcinoma.Front Immunol. 2024 Aug 13;15:1440764. doi: 10.3389/fimmu.2024.1440764. eCollection 2024. Front Immunol. 2024. PMID: 39192980 Free PMC article. Review.
-
Natural killer cell dysfunction is associated with colorectal cancer with severe COVID-19.World J Gastrointest Oncol. 2025 May 15;17(5):104591. doi: 10.4251/wjgo.v17.i5.104591. World J Gastrointest Oncol. 2025. PMID: 40487965 Free PMC article.
-
Antibody-dependent enhancement of coronaviruses.Int J Biol Sci. 2025 Feb 3;21(4):1686-1704. doi: 10.7150/ijbs.96112. eCollection 2025. Int J Biol Sci. 2025. PMID: 39990674 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

