Serum Lipidome Profiling Reveals a Distinct Signature of Ovarian Cancer in Korean Women
- PMID: 38412029
- PMCID: PMC11061607
- DOI: 10.1158/1055-9965.EPI-23-1293
Serum Lipidome Profiling Reveals a Distinct Signature of Ovarian Cancer in Korean Women
Abstract
Background: Distinguishing ovarian cancer from other gynecological malignancies is crucial for patient survival yet hindered by non-specific symptoms and limited understanding of ovarian cancer pathogenesis. Accumulating evidence suggests a link between ovarian cancer and deregulated lipid metabolism. Most studies have small sample sizes, especially for early-stage cases, and lack racial/ethnic diversity, necessitating more inclusive research for improved ovarian cancer diagnosis and prevention.
Methods: Here, we profiled the serum lipidome of 208 ovarian cancer, including 93 early-stage patients with ovarian cancer and 117 nonovarian cancer (other gynecological malignancies) patients of Korean descent. Serum samples were analyzed with a high-coverage liquid chromatography high-resolution mass spectrometry platform, and lipidome alterations were investigated via statistical and machine learning (ML) approaches.
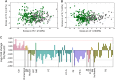
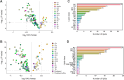
Results: We found that lipidome alterations unique to ovarian cancer were present in Korean women as early as when the cancer is localized, and those changes increase in magnitude as the diseases progresses. Analysis of relative lipid abundances revealed specific patterns for various lipid classes, with most classes showing decreased abundance in ovarian cancer in comparison with other gynecological diseases. ML methods selected a panel of 17 lipids that discriminated ovarian cancer from nonovarian cancer cases with an AUC value of 0.85 for an independent test set.
Conclusions: This study provides a systemic analysis of lipidome alterations in human ovarian cancer, specifically in Korean women.
Impact: Here, we show the potential of circulating lipids in distinguishing ovarian cancer from nonovarian cancer conditions.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Mercado C, Zingmond D, Karlan BY, Sekaris E, Gross J, Maggard-Gibbons M, et al. Quality of care in advanced ovarian cancer: the importance of provider specialty. Gynecol Oncol 2010;117:18–22. - PubMed
-
- Kobayashi E, Ueda Y, Matsuzaki S, Yokoyama T, Kimura T, Yoshino K, et al. Biomarkers for screening, diagnosis, and monitoring of ovarian cancer. Cancer Epidemiol Biomarkers Prev 2012;21:1902–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

