A VRC13-like bNAb response is associated with complex escape pathways in HIV-1 envelope
- PMID: 38412036
- PMCID: PMC10949433
- DOI: 10.1128/jvi.01720-23
A VRC13-like bNAb response is associated with complex escape pathways in HIV-1 envelope
Abstract
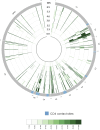
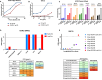
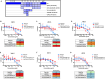
The rational design of HIV-1 immunogens to trigger the development of broadly neutralizing antibodies (bNAbs) requires understanding the viral evolutionary pathways influencing this process. An acute HIV-1-infected individual exhibiting >50% plasma neutralization breadth developed neutralizing antibody specificities against the CD4-binding site (CD4bs) and V1V2 regions of Env gp120. Comparison of pseudoviruses derived from early and late autologous env sequences demonstrated the development of >2 log resistance to VRC13 but not to other CD4bs-specific bNAbs. Mapping studies indicated that the V3 and CD4-binding loops of Env gp120 contributed significantly to developing resistance to the autologous neutralizing response and that the CD4-binding loop (CD4BL) specifically was responsible for the developing resistance to VRC13. Tracking viral evolution during the development of this cross-neutralizing CD4bs response identified amino acid substitutions arising at only 4 of 11 known VRC13 contact sites (K282, T283, K421, and V471). However, each of these mutations was external to the V3 and CD4BL regions conferring resistance to VRC13 and was transient in nature. Rather, complete resistance to VRC13 was achieved through the cooperative expression of a cluster of single amino acid changes within and immediately adjacent to the CD4BL, including a T359I substitution, exchange of a potential N-linked glycosylation (PNLG) site to residue S362 from N363, and a P369L substitution. Collectively, our data characterize complex HIV-1 env evolution in an individual developing resistance to a VRC13-like neutralizing antibody response and identify novel VRC13-associated escape mutations that may be important to inducing VRC13-like bNAbs for lineage-based immunogens.IMPORTANCEThe pursuit of eliciting broadly neutralizing antibodies (bNAbs) through vaccination and their use as therapeutics remains a significant focus in the effort to eradicate HIV-1. Key to our understanding of this approach is a more extensive understanding of bNAb contact sites and susceptible escape mutations in HIV-1 envelope (env). We identified a broad neutralizer exhibiting VRC13-like responses, a non-germline restricted class of CD4-binding site antibody distinct from the well-studied VRC01-class. Through longitudinal envelope sequencing and Env-pseudotyped neutralization assays, we characterized a complex escape pathway requiring the cooperative evolution of four amino acid changes to confer complete resistance to VRC13. This suggests that VRC13-class bNAbs may be refractory to rapid escape and attractive for therapeutic applications. Furthermore, the identification of longitudinal viral changes concomitant with the development of neutralization breadth may help identify the viral intermediates needed for the maturation of VRC13-like responses and the design of lineage-based immunogens.
Keywords: CD4bs; HIV; VRC13; bNAb; escape; evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Li B, Decker JM, Johnson RW, Bibollet-Ruche F, Wei X, Mulenga J, Allen S, Hunter E, Hahn BH, Shaw GM, Blackwell JL, Derdeyn CA. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol 80:5211–5218. doi: 10.1128/JVI.00201-06 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

