Filamin B restricts vaccinia virus spread and is targeted by vaccinia virus protein C4
- PMID: 38412044
- PMCID: PMC10949515
- DOI: 10.1128/jvi.01485-23
Filamin B restricts vaccinia virus spread and is targeted by vaccinia virus protein C4
Abstract
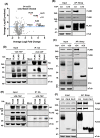
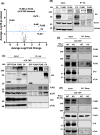
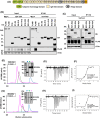
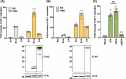
Vaccinia virus (VACV) is a large DNA virus that encodes scores of proteins that modulate the host immune response. VACV protein C4 is one such immunomodulator known to inhibit the activation of both the NF-κB signaling cascade and the DNA-PK-mediated DNA sensing pathway. Here, we show that the N-terminal region of C4, which neither inhibits NF-κB nor mediates interaction with DNA-PK, still contributes to virus virulence. Furthermore, this domain interacts directly and with high affinity to the C-terminal domain of filamin B (FLNB). FLNB is a large actin-binding protein that stabilizes the F-actin network and is implicated in other cellular processes. Deletion of FLNB from cells results in larger VACV plaques and increased infectious viral yield, indicating that FLNB restricts VACV spread. These data demonstrate that C4 has a new function that contributes to virulence and engages the cytoskeleton. Furthermore, we show that the cytoskeleton performs further previously uncharacterized functions during VACV infection.
Importance: Vaccinia virus (VACV), the vaccine against smallpox and monkeypox, encodes many proteins to counteract the host immune response. Investigating these proteins provides insights into viral immune evasion mechanisms and thereby indicates how to engineer safer and more immunogenic VACV-based vaccines. Here, we report that the N-terminal domain of VACV protein C4 interacts directly with the cytoskeletal protein filamin B (FLNB), and this domain of C4 contributes to virus virulence. Furthermore, VACV replicates and spreads better in cells lacking FLNB, thus demonstrating that FLNB has antiviral activity. VACV utilizes the cytoskeleton for movement within and between cells; however, previous studies show no involvement of C4 in VACV replication or spread. Thus, C4 associates with FLNB for a different reason, suggesting that the cytoskeleton has further uncharacterized roles during virus infection.
Keywords: DNA sensing; NF-κB; cytoskeleton; filamin B; protein C4; vaccinia virus; virus restriction factor; virus virulence; virus-host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Moss B, Smith GL. 2021. Poxviridae: the viruses and their replication, p 573–613. In Howley PM, Knipe DM (ed), Fields virology: DNA viruses, 7th ed, Vol. 2. Wolters Kluwer Inc.
-
- Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID. 1988. Smallpox and its eradication. World Health Organization, Geneva.
-
- Panicali D, Davis SW, Weinberg RL, Paoletti E. 1983. Construction of live vaccines by using genetically engineered poxviruses: biological activity of recombinant vaccinia virus expressing influenza virus hemagglutinin. Proc Natl Acad Sci U S A 80:5364–5368. doi: 10.1073/pnas.80.17.5364 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

