A billion years of evolution manifest in nanosecond protein dynamics
- PMID: 38412135
- PMCID: PMC10927572
- DOI: 10.1073/pnas.2318743121
A billion years of evolution manifest in nanosecond protein dynamics
Abstract
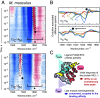
Protein dynamics form a critical bridge between protein structure and function, yet the impact of evolution on ultrafast processes inside proteins remains enigmatic. This study delves deep into nanosecond-scale protein dynamics of a structurally and functionally conserved protein across species separated by almost a billion years, investigating ten homologs in complex with their ligand. By inducing a photo-triggered destabilization of the ligand inside the binding pocket, we resolved distinct kinetic footprints for each homolog via transient infrared spectroscopy. Strikingly, we found a cascade of rearrangements within the protein complex which manifest in time points of increased dynamic activity conserved over hundreds of millions of years within a narrow window. Among these processes, one displays a subtle temporal shift correlating with evolutionary divergence, suggesting reduced selective pressure in the past. Our study not only uncovers the impact of evolution on molecular processes in a specific case, but has also the potential to initiate a field of scientific inquiry within molecular paleontology, where species are compared and classified based on the rapid pace of protein dynamic processes; a field which connects the shortest conceivable time scale in living matter (10[Formula: see text] s) with the largest ones (10[Formula: see text] s).
Keywords: biophysics; evolution; photoswitch; protein dynamics; transient infrared spectroscopy.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Similar articles
-
Planning Implications Related to Sterilization-Sensitive Science Investigations Associated with Mars Sample Return (MSR).Astrobiology. 2022 Jun;22(S1):S112-S164. doi: 10.1089/AST.2021.0113. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904892
-
Predicting ligand binding affinity using on- and off-rates for the SAMPL6 SAMPLing challenge.J Comput Aided Mol Des. 2018 Oct;32(10):1001-1012. doi: 10.1007/s10822-018-0149-3. Epub 2018 Aug 23. J Comput Aided Mol Des. 2018. PMID: 30141102 Free PMC article.
-
Heterogeneity and dynamics of the ligand recognition mode in purine-sensing riboswitches.Biochemistry. 2010 May 4;49(17):3703-14. doi: 10.1021/bi1000036. Biochemistry. 2010. PMID: 20345178
-
Ligand diffusion in proteins via enhanced sampling in molecular dynamics.Phys Life Rev. 2017 Dec;22-23:58-74. doi: 10.1016/j.plrev.2017.03.003. Epub 2017 Apr 1. Phys Life Rev. 2017. PMID: 28410930 Review.
-
Biophysical Models of Protein Evolution: Understanding the Patterns of Evolutionary Sequence Divergence.Annu Rev Biophys. 2017 May 22;46:85-103. doi: 10.1146/annurev-biophys-070816-033819. Epub 2017 Mar 15. Annu Rev Biophys. 2017. PMID: 28301766 Free PMC article. Review.
References
-
- Jankovic B., et al. , Sequence of events during peptide unbinding from RNase S: A complete experimental description. J. Phys. Chem. Lett. 12, 5201–5207 (2021). - PubMed
-
- Heckmeier P. J., Ruf J., Buhrke D., Janković B. G., Hamm P., Signal propagation within the MCL-1/BIM protein complex. J. Mol. Biol. 434, 167499 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

