The TaSnRK1-TabHLH489 module integrates brassinosteroid and sugar signalling to regulate the grain length in bread wheat
- PMID: 38412139
- PMCID: PMC11182588
- DOI: 10.1111/pbi.14319
The TaSnRK1-TabHLH489 module integrates brassinosteroid and sugar signalling to regulate the grain length in bread wheat
Abstract
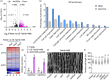
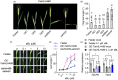
Regulation of grain size is a crucial strategy for improving the crop yield and is also a fundamental aspect of developmental biology. However, the underlying molecular mechanisms governing grain development in wheat remain largely unknown. In this study, we identified a wheat atypical basic helix-loop-helix (bHLH) transcription factor, TabHLH489, which is tightly associated with grain length through genome-wide association study and map-based cloning. Knockout of TabHLH489 and its homologous genes resulted in increased grain length and weight, whereas the overexpression led to decreased grain length and weight. TaSnRK1α1, the α-catalytic subunit of plant energy sensor SnRK1, interacted with and phosphorylated TabHLH489 to induce its degradation, thereby promoting wheat grain development. Sugar treatment induced TaSnRK1α1 protein accumulation while reducing TabHLH489 protein levels. Moreover, brassinosteroid (BR) promotes grain development by decreasing TabHLH489 expression through the transcription factor BRASSINAZOLE RESISTANT1 (BZR1). Importantly, natural variations in the promoter region of TabHLH489 affect the TaBZR1 binding ability, thereby influencing TabHLH489 expression. Taken together, our findings reveal that the TaSnRK1α1-TabHLH489 regulatory module integrates BR and sugar signalling to regulate grain length, presenting potential targets for enhancing grain size in wheat.
Keywords: SnRK1; TabHLH489; brassinosteroid; sugar; wheat grain length.
© 2024 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no competing interests.
Figures







Similar articles
-
TabHLH489 suppresses nitrate signaling by inhibiting the function of TaNLP7-3A in wheat.J Integr Plant Biol. 2025 Apr;67(4):1162-1178. doi: 10.1111/jipb.13832. Epub 2024 Dec 29. J Integr Plant Biol. 2025. PMID: 39737613
-
The brassinosteroid signaling-related ILI-OsAIF-OsbHLH92 transcription factor module antagonistically controls leaf angle and grain size in rice.Plant Physiol. 2024 Dec 24;197(1):kiae668. doi: 10.1093/plphys/kiae668. Plant Physiol. 2024. PMID: 39704312
-
The basic helix-loop-helix transcription factor OsBLR1 regulates leaf angle in rice via brassinosteroid signalling.Plant Mol Biol. 2020 Apr;102(6):589-602. doi: 10.1007/s11103-020-00965-5. Epub 2020 Feb 5. Plant Mol Biol. 2020. PMID: 32026326
-
TabHLH95-TaNF-YB1 module promotes grain starch synthesis in bread wheat.J Genet Genomics. 2023 Nov;50(11):883-894. doi: 10.1016/j.jgg.2023.04.002. Epub 2023 Apr 14. J Genet Genomics. 2023. PMID: 37062449
-
Brassinosteroid-mediated regulation of agronomic traits in rice.Plant Cell Rep. 2014 May;33(5):683-96. doi: 10.1007/s00299-014-1578-7. Epub 2014 Mar 26. Plant Cell Rep. 2014. PMID: 24667992 Free PMC article. Review.
Cited by
-
GmGIF5 Promotes Cell Expansion by Negatively Regulating Cell Wall Modification.Int J Mol Sci. 2025 Jan 9;26(2):492. doi: 10.3390/ijms26020492. Int J Mol Sci. 2025. PMID: 39859208 Free PMC article.
-
The transcription factor CAMTA2 interacts with the histone acetyltransferase GCN5 and regulates grain weight in wheat.Plant Cell. 2024 Sep 25;36(12):4895-913. doi: 10.1093/plcell/koae261. Online ahead of print. Plant Cell. 2024. PMID: 39321218 Free PMC article.
-
Induced variation in BRASSINOSTEROID INSENSITIVE 1 (BRI1) confers a compact wheat architecture.BMC Plant Biol. 2025 May 26;25(1):700. doi: 10.1186/s12870-025-06762-w. BMC Plant Biol. 2025. PMID: 40419954 Free PMC article.
-
Somatic embryogenetic receptor kinase TaSERL2 regulates heat stress tolerance in wheat by influencing TaBZR2 protein stability and transcriptional activity.Plant Biotechnol J. 2025 Jul;23(7):2537-2553. doi: 10.1111/pbi.70045. Epub 2025 Apr 4. Plant Biotechnol J. 2025. PMID: 40183279 Free PMC article.
-
Advances in genome editing in plants within an evolving regulatory landscape, with a focus on its application in wheat breeding.J Plant Biochem Biotechnol. 2025;34(3):599-614. doi: 10.1007/s13562-025-00981-w. Epub 2025 Apr 15. J Plant Biochem Biotechnol. 2025. PMID: 40852733 Free PMC article. Review.
References
-
- Barrett, J.C. , Fry, B. , Maller, J. and Daly, M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. - PubMed
-
- Che, R. , Tong, H. , Shi, B. , Liu, Y. , Fang, S. , Liu, D. , Xiao, Y. et al. (2015) Control of grain size and rice yield by GL2‐mediated brassinosteroid responses. Nat. Plants, 2, 15195. - PubMed
MeSH terms
Substances
Grants and funding
- 2021YFD1201500/The National Key Research and Development Program of China
- 31970306/The National Natural Science Foundation of China
- 32070210/The National Natural Science Foundation of China
- 32100267/The National Natural Science Foundation of China
- 32100492/The National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

